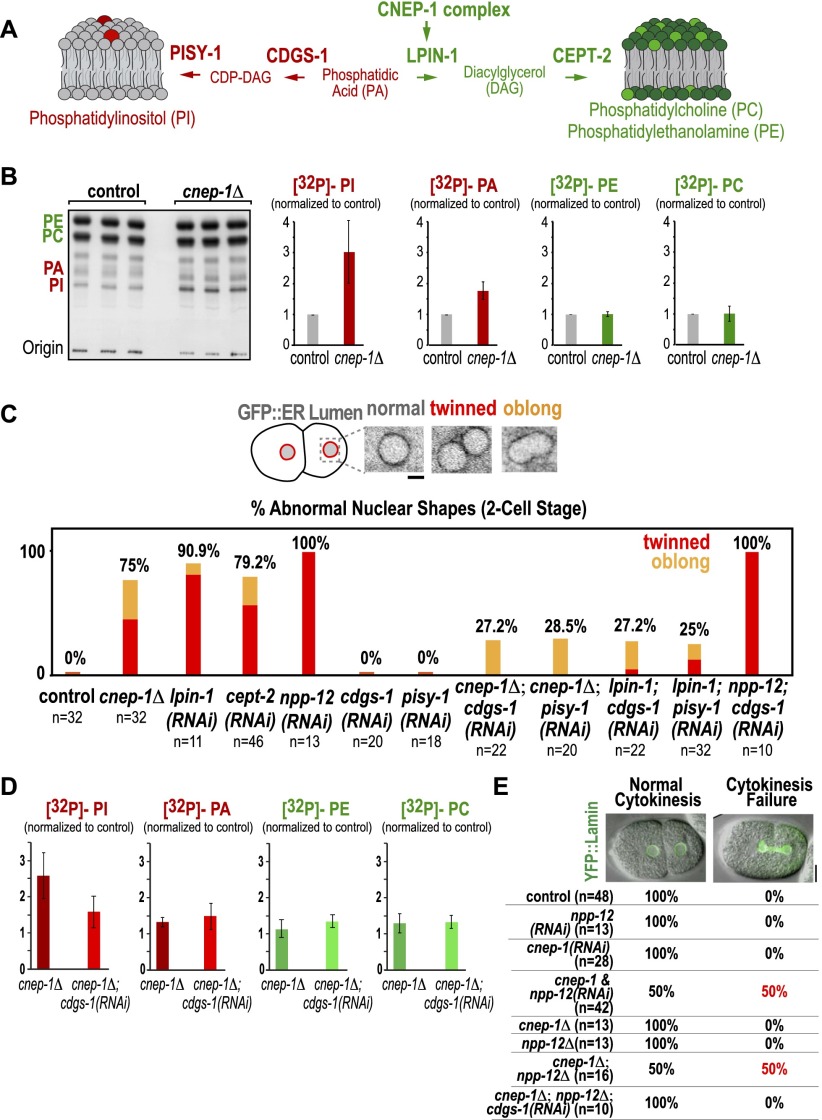
Figure 3.
The nuclear envelope disassembly defect in the cnep-1Δ mutant results from increased PI synthesis. (A) Schematic highlighting the C. elegans enzymes required to synthesize PI from the lipin substrate PA (CDGS-1 and PISY-1; red) and to synthesize PC and PE from the lipin product DAG (CEPT-2; green). (B) Autoradiograph of a chromatography plate after separation of isolated 32P-labeled phospholipids. Graphs plotting the mean band intensity for each phospholipid species from three independent replicates normalized against the mean value in control samples run in parallel. Error bars are the SD. (C, top) Inverted contrast confocal images of representative nuclear phenotypes in two-cell stage embryos expressing the ER lumen marker GFP∷SP-12. (Bottom) Plots quantifying abnormal nuclear phenotypes at the two-cell stage for the indicated conditions. Bar, 2 µm. (D) Graphs plotting the mean band intensity for each phospholipid species from three independent replicates normalized against the mean value in control samples run in parallel. Error bars are the SD. (E, top) Overlay of differential interference contrast and YFP∷lamin fluorescence confocal images for a control embryo (normal cytokinesis) and a cnep-1 and npp-12(RNAi) embryo failing cytokinesis (cytokinesis failure). (Bottom) Table showing the percentage of embryos exhibiting cytokinesis success or failure.