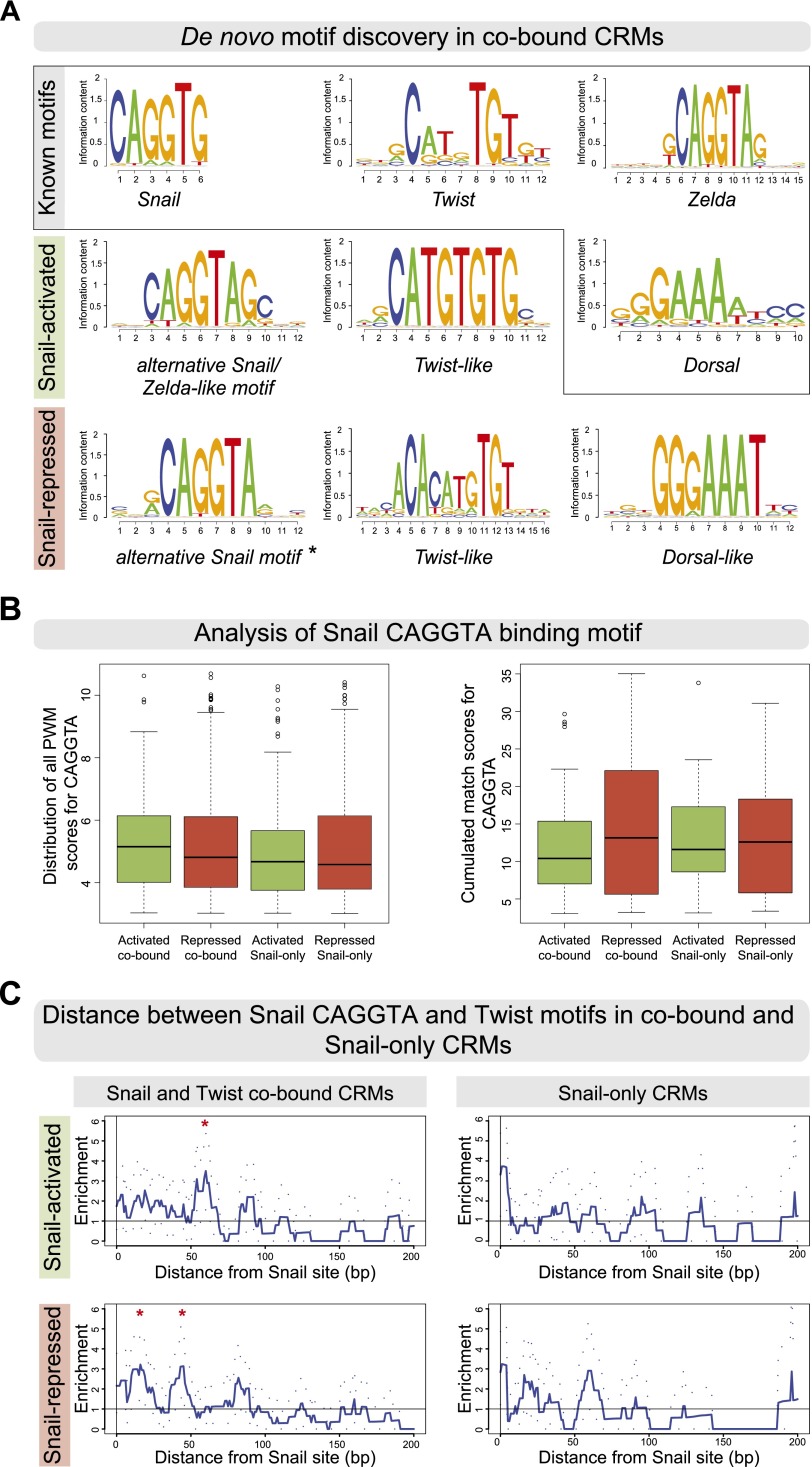
Figure 4.
Snail-activated and -repressed enhancers contain subtle differences in their Snail and Twist motifs. (A) De novo motif discovery in cobound activated and repressed CRMs. Position weight matrices (PWMs) are shown as sequence logos, with known motifs for Snail (Jaspar MA0086.1), Twist (FlyReg), Zelda (SOLEXA_5), and Dorsal (FlyReg). An alternative Snail and a Twist-like motif are found in both sets, while only repressed CRMs are enriched for a Dorsal-like motif. (*) Alternative Snail motif used for analysis shown in B and C. (B) Distribution of PWM match scores (P-value < 1 × 10−3) for the alternative CAGGTA motif across the four classes of Snail-bound CRMs showing all PWM matches (Patser scores, left box plot) or the cumulated match scores (right box plot), summing up putative high-affinity and low-affinity sites. In both cases, no significant differences in the number of motifs were observed. (C) The base-pair distance between Twist and CAGGTA Snail motifs is greater in activated compared with repressed cobound CRMs. The Y-axis shows the mean enrichment of Snail–Twist distances over random expectations (smoothed using a 10-bp window), with 95% confidence intervals (dotted lines). Red asterisks indicate where the signal deviates from random (confidence interval remains less than one for greater than five consecutive values). (Top panel) In activated cobound CRMs, Twist motifs are preferentially enriched at a distance of 50–65 bp. (Bottom panel) In repressed cobound CRMs, Twist motifs cluster around Snail motifs at a distance of 10–20 and 40–50 bp. No enrichment of Twist motifs around CAGGTA Snail motifs is seen in Snail-only CRMs, as expected.