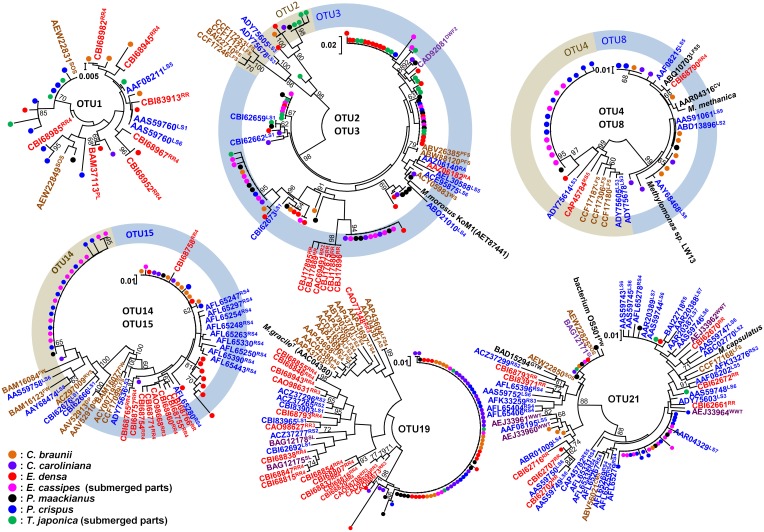
Figure 5.
Neighbor-joining tree of individual clones in abundant OTUs and their related environmental clones based on deduced amino acid sequences of pmoA genes. The clones obtained in this study were indicated as closed circles. Bootstrap values (>60%) obtained from 1000 resamplings are shown. Related clones are indicated as accession numbers with superscript indicating their origins as follows; plants in red including RR, rice root and PL, plant leaf, saturated soil or sediment in blue including RS, rice field soil; LS, lake sediment; RA, river aquifer; TCE, TCE-contaminated aquifer; WS, wetland soil; and RVS, river sediment, unsaturated soil in brown including AS, agricultural soil; PFS, permafrost soil; LFS, landfill-cover soil; SOS, surface soils of onshore oil and gas fields; and CMS, coal mine soil, clone from water in purple including DWF, drinking water facility; SL, methane-consuming sludge; WWT, wastewater treatment plant; DP, drainage of peat land; GTW, subsurface geothermal water stream in a Japanese gold mine, and clones from biomat in black including FWI, freshwater iron-rich microbial mat; PW, pond water; and CV, biomat and water in cave.