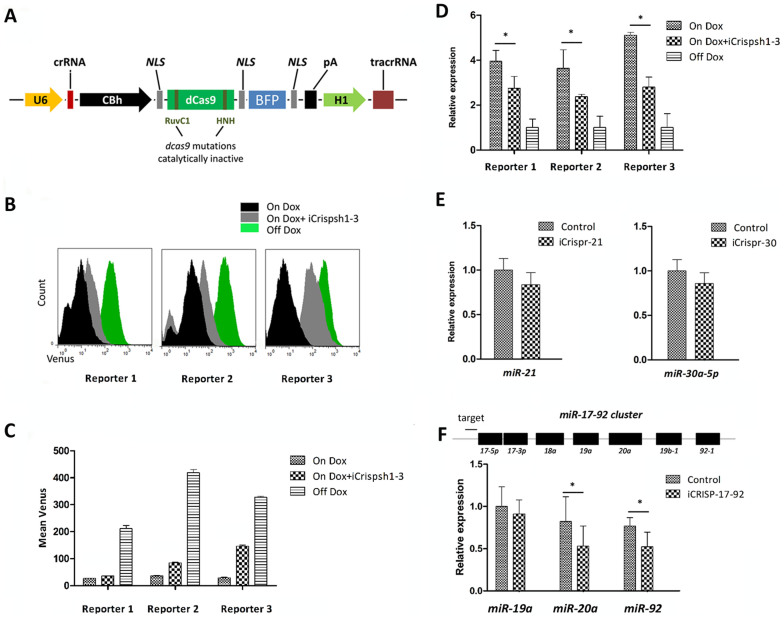
Figure 2. Repression of shRNAs and miRNAs using CRISPRi in NIH3T3 cells.
(A). The CRISPRi system consists of a fusion protein and two designable RNA elements. The dCas9 protein is defective for nuclease activity but can block RNA polymerase binding and transcriptional elongation when targeting occurs. All crRNA sequences were identical to those described in Table S1. (B). FCM of reporter cells transfected with iCrispsh1, iCrispsh 2 or iCrispsh3 carrying dCas9/gRNA cassettes, with or without Dox treatment (On/Off Dox). The fluorescence intensity distribution after 72 h indicated significant repression of the shRNAs by CRISPRi. (C). Relative analysis histograms were generated from FCM. The inhibition effect of CRISPRi was 40–50% for each shRNA. The values shown are the means of three replicates. (D). Examination of the reads per shRNA indicated a strong repressive effect on miRNAs (10–30% fold) as assayed using quantitative PCR. The values shown are the means of three replicates. (E). NIH3T3 cells were transfected with iCrispr-21 and iCrispr-30. The qPCR data show that transfection of repressing vector can inhibit endogenous miR-21 and miR-30a miRNA expression. The values shown are the means of three replicates, and each miRNA was normalized to an internal control, U6 RNA. (F). The miR-17-92 cluster encodes 7 miRNAs. A crRNA targeting an area upstream of the cluster was designed for use with CRISPRi. We hypothesized that iCrisp-17-92 was able to repress all miRNA expression within the entire cluster by transcriptional elongation blockage. qPCR was performed in three replicates, there was a repressive effect on miR-19a expression (10% fold) and significant repression of miR-20a and miR-92-1 expression was observed (20–25% fold). The values shown are the means of three replicates. Single star indicated P < 0.05.