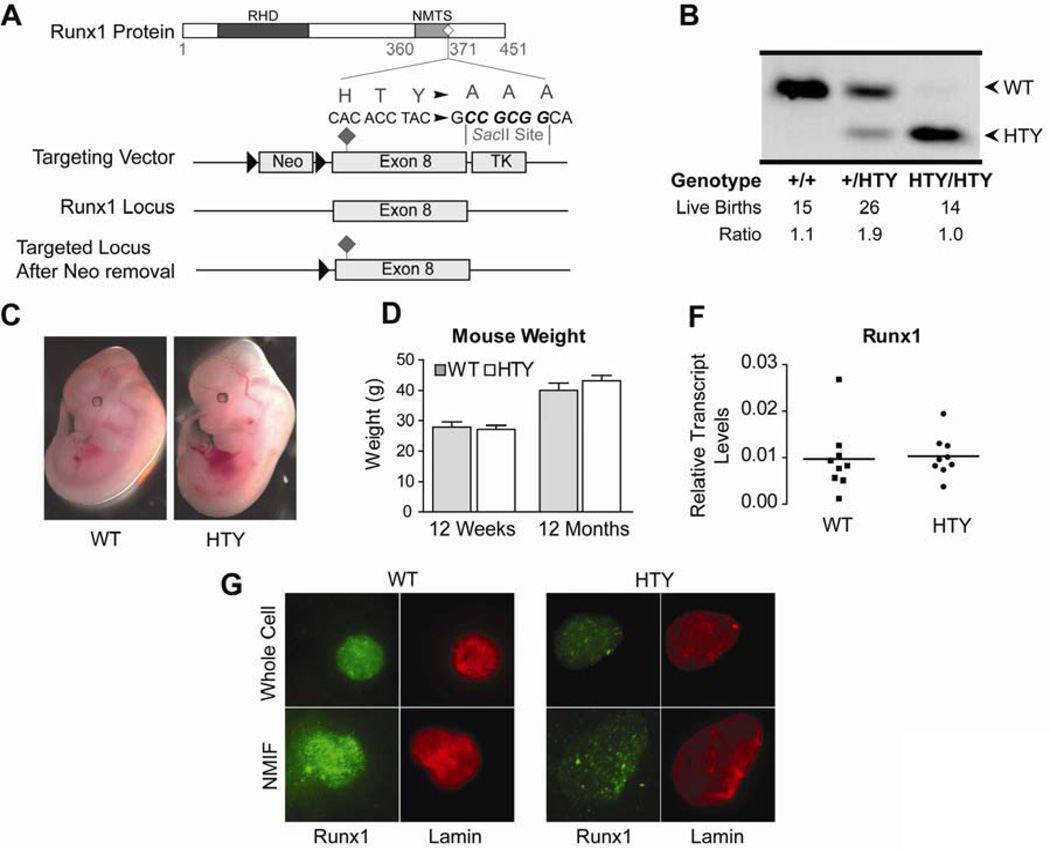
Figure 1. Generation of the Runx1HTY350-352AAA mouse.
(A) Targeting vector for the knock-in mouse contained a floxed Neo cassette as a positive selection marker and a TK cassette for negative selection to ensure homologous recombination in ES cells. (B) Successful targeting to the native Runx1 locus results in a diagnostic SacII site that allows for genotyping by PCR and enzyme digest. From left to right are genotyping results showing the change in digest fragment sizes from a wildtype, heterozygous and homozygous Runx1HTY350-352AAA mouse. (C) Runx1HTY350-352AAA animals were born at Mendelian ratios. (D) Runx1HTY350-352AAA embryos are healthy at embryonic day 12.5 and are comparable to wildtype littermates (3 litters, n=8 WT, 7 HTY). (E) Adult wildtype and Runx1HTY350-352AAA mice at 12 weeks or 12 months are equivalent in weight (n=16 WT, 19 HTY for 12 weeks and 15 WT, 9 HTY for 12 months). (F) qRT-PCR for Runx1 in bone marrow cells shows no difference in expression levels (p=0.8 by Student’s T test). Each point represents the average of technical replicates from one animal, normalized to mCox (n=9 WT, 9 HTY). (G) Whole cell and nuclear matrix-intermediate filament (NMIF) preparations of bone marrow from wildtype and Runx1HTY350-352AAA animals shows that Runx1 is still present within the nuclear matrix (n=3 WT, 3 HTY). All of the images were acquired using same settings of the microscope and the associated MetaMorph Software for accurate comparison. All error bars are SEM.