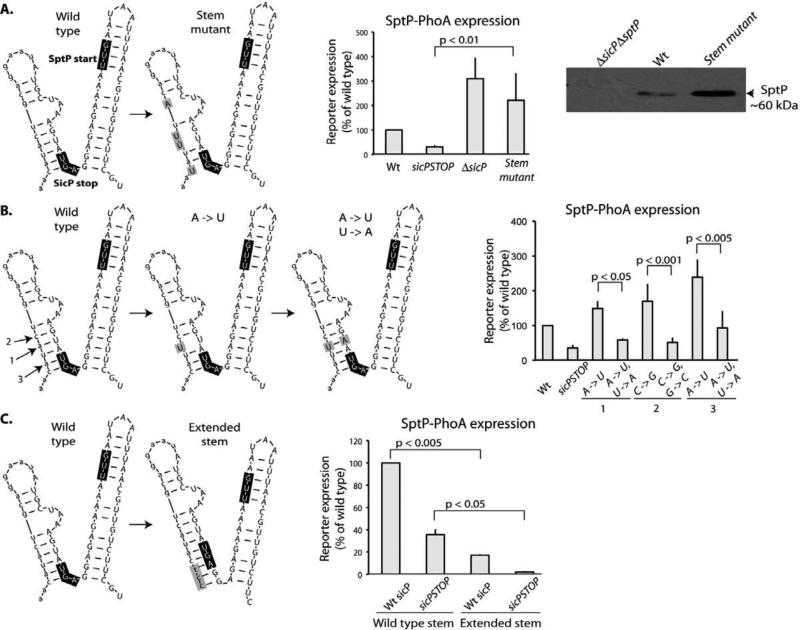
Fig. 4.
An mRNA structure inhibits translation of sptP. (A) The mutations indicated in the schematic representation (in grey boxes) of secondary mRNA structure surrounding the sptP start codon (as predicted by mFOLD) were introduced into a strain encoding a sptP::phoA reporter and whole cell lysates were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Additionally, the same mutations were introduced into a wild type strain and whole cell lysates were analyzed by western immunoblot with an anti-SptP antibody. (B) The mutations indicated in the schematic representation (in grey boxes) predicted to disrupt (A -> U) or restore (A -> U/U-> A) base complementarity in the predicted stem surrounding the sptP start codon were introduced into a background strain encoding a sptP::phoA reporter. Whole cell lysates of these strains were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Student's t test was used to determine p values. (C) The indicated mutations to lengthen the predicted stem surrounding the sptP start codon were introduced into a background strain encoding a sptP::phoA reporter. Whole cell lysates of these strains were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Student's t test was used to determine p values.