Figure 1.
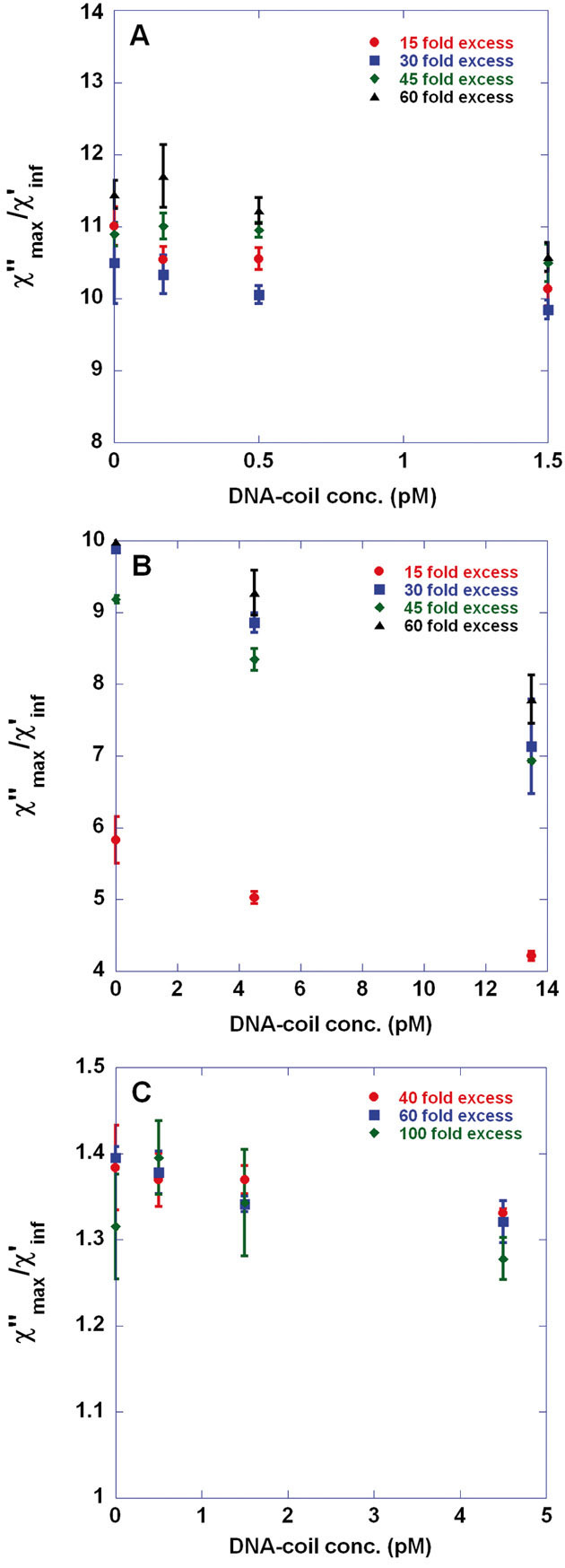
Singleplex detection of V. cholerae (VC) target-DNA by means of applying the volume-amplified magnetic nanobead detection assay using 100 and 250 nm magnetic nanobeads. Biodetection procedure comprised the steps of (i) target recognition through padlock probe assay giving DNA-circles; (ii) 1 h rolling circle amplification of the DNA-circles giving DNA-coils and (iii) immobilization of detection oligonucleotide-tagged magnetic nanobeads exhibiting Brownian relaxation behavior in the DNA-coils. Detection oligonucleotides were conjugated to the beads using avidin-biotin chemistry. By recording the frequency-dependent magnetic susceptibility of the sample, χ = χ‘ – iχ“, using a portable AC susceptometer (DynoMag®) target quantification could be obtained by considering the amplitude of the Brownian relaxation peak for non-immobilized nanobeads; χ“max. (A, B) χ“max versus VC DNA-coil concentration (0–1.5 and 0–13.5 pM in panels A and B, respectively) recorded at 24°C for 100 nm beads conjugated with 15-, 30-, 45-, and 60-fold excess of detection oligonucleotides with respect to the number of beads. (C) χ“max versus VC DNA-coil concentration (0–4.5 pM) recorded at 24°C for 250 nm beads conjugated with 40-, 60-, and 100-fold excess of detection oligonucleotides with respect to the number of beads. To account for differences in nanobead content between samples, the χ“max data have been normalized using the constant high-frequency χ‘ level, χ‘inf. The error bars show one standard deviation based on three independent measurements.
