Figure 2.
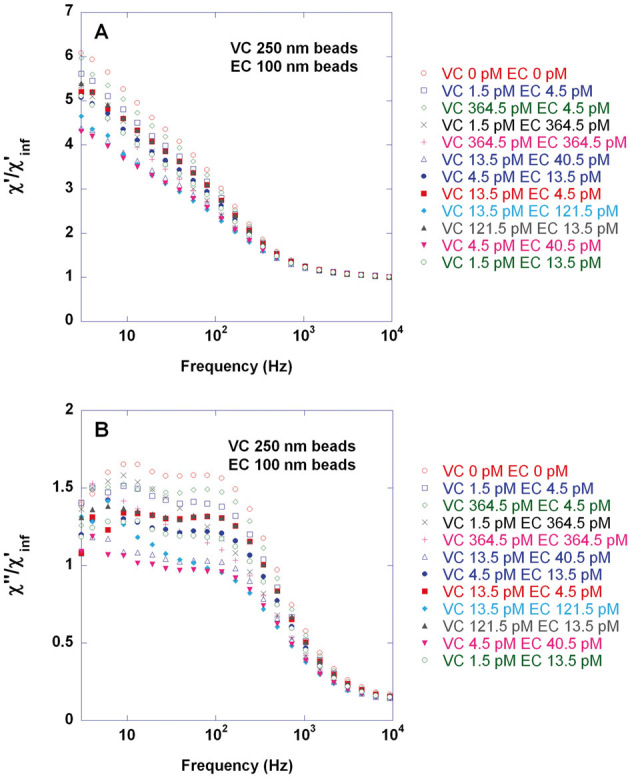
Biplex detection of V. cholerae (VC) and E. coli (EC) target-DNA for twelve combinations of target concentrations by means of the volume-amplified magnetic nanobead detection assay and using 250 nm and 100 nm magnetic nanobeads, respectively. Biodetection procedure comprised the steps of (i) target recognition performed separately for VC and EC through padlock probe assay giving DNA-circles; (ii) 1 hr rolling circle amplification of the mixtures of VC and EC DNA-circles giving a series of samples containing different combinations of concentrations of VC and EC DNA-coils and (iii) iimmobilization of a mixture of 250 nm and 100 nm magnetic nanobeads exhibiting Brownian relaxation behavior and conjugated with VC and EC detection oligonucleotides using avidin-biotin chemistry. EC and VC detection probes were conjugated to 100 nm and 250 nm beads using a 30 and 60 fold excess of oligonucleotides (found to give optimal VC singleplex detection sensitivity), respectively. Target quantification could be obtained by recording the frequency-dependent magnetic susceptibility of the sample, χ = χ‘ – iχ“, using a portable AC susceptometer (DynoMag®) followed by fitting each χ“ curve to a double Cole–Cole expression (sum of two Cole–Cole distributions) in order to extract χ“max (HFP level) for VC and EC. The figure shows real (A) and imaginary (B) parts of the magnetic susceptibility recorded at 24°C and normalized using the constant high-frequency χ‘ level, χ‘inf. Each concentration combination was measured once.
