Abstract
This study aimed to isolate and identify Lactobacillus in the honey stomach of honeybee Apis dorsata. Samples of honeybee were collected from A. dorsata colonies in different bee trees and Lactobacillus bacteria isolated from honey stomachs. Ninety two isolates were Gram-stained and tested for catalase reaction. By using bacterial universal primers, the 16S rDNA gene from DNA of bacterial colonies amplified with polymerase chain reaction (PCR). Forty-nine bacterial 16S rDNA gene were sequenced and entrusted in GenBank. Phylogenetic analysis showed they were different phylotypes of Lactobacillus. Two of them were most closely relevant to the previously described species Lactobacillus plantarum. Other two phylotypes were identified to be closely related to Lactobacillus pentosus. However, only one phylotype was found to be distantly linked to the Lactobacillus fermentum. The outcomes of the present study indicated that L. plantarum, L. pentosus, and L. fermentum were the dominant lactobacilli in the honey stomach of honeybee A. dorsata collected during the dry season from Malaysia forest area - specifically “Melaleuca in Terengganu”.
Keywords: honey bee, honey stomach, Lactobacillus, probiotic, 16S rDNA gene sequences
Introduction
The genus Lactobacillus is one of the most important genera of lactic acid bacteria (LAB) and includes over 174 species (Dsmz, 2008). Lactobacillus bacteria are catalase negative, Gram positive and usually rod shaped. They are found in a wide variety of habitats, including vegetable and the gastrointestinal tracts of animals, like honeybees (Tannock, 2004; Schrezenmeir and de Vrese, 2001; Fujisawa and Mitsuoka, 1996) and are employed in manufacturing of fermented foods, such as soybean products, fish products, vegetable products, bread, alcoholic beverages, dairy, and for cheese production (Kandler et al., 1986; Rhee et al., 2011; Caplice and Fitzgerald, 1999). The dominant LAB species in the honey stomach of honeybees’ A. mellifera, A. dorsata, and bumblebees have been reported to belong to the Lactobacillus genus (Olofsson and Vasquez, 2008; Vásquez et al., 2009; Tajabadi et al., 2011b).
Microorganisms present in honey are those surviving the low pH and antimicrobial compounds found in honey. The microorganisms probably originate from pollen, the digestive tracts of honeybees, flowers, dirt, dust, and the air. Gilliam (1997) discovered that honeycomb was dominated mainly by bacteria and yeast, which all arrived from the bees or the raw materials (nectar) (Gilliam, 1997). Larvae could be sterile initially, but since they eat pollens and nectar supplied by the workers, hence they are inoculated prior to pupation (Lee and Kime, 1984).
The distinguishable phenotypic property for detection and identification of LAB microbiota has been leading to a trend towards using molecular-based identification methods. Different approaches of rDNA gene sequencing (Wallbanks et al., 1990; Rachman et al., 2003; Settanni et al., 2005; Yoshiyama and Kimura, 2009; Snell-Castro et al., 2005) have been employed to identify Lactobacillus species. Within the recent years, sequencing of 16S rDNA and its use has been considered as the “gold standard” for identification and phylogenetic analysis of LAB (Ludwig and Schleifer, 1999).
The use of Lactobacillus species as probiotics has been found to enhance the immunity of honeybees, helping them to survive against the effect of pathogens and bring advantageous properties for honeybee health (Evans and Lopez, 2004; Forsgren et al., 2010). Therefore, the present study aimed to isolate and identify lactobacilli strains from the honey stomach of honeybees A. dorsata collected during the dry season from Malaysia forest area - specifically “Melaleuca in Terengganu”.
Materials and Methods
Sampling
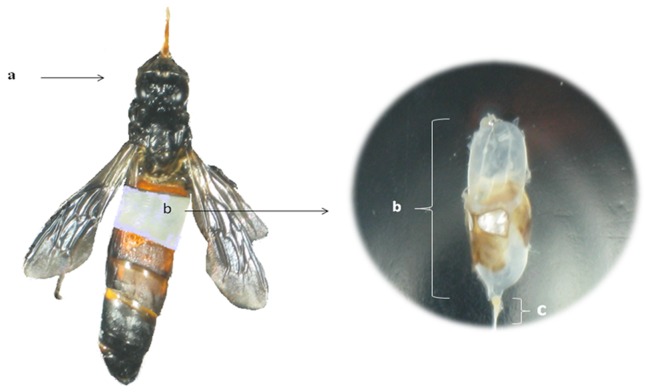
During honey hunting, ten samples of bees were collected from A. dorsata wild colonies over the course of dry season between June and August 2009 from various bee trees in the secondary forest area of Melaleuca, in Terengganu, Malaysia. Samples were placed in individual 20 mL sterile tubes containing 10 mL of sterile 0.1% peptone saline solution supplemented with Tween 80 (0.9% w/v NaCl- Merck, Darmstadt, Germany -, 0.1% w/v Tween 80 -Merck, Darmstadt, Germany -, and 0.1% w/v Peptone -Merck, Darmstadt, Germany) and immediately transported to the Food Biotechnology Laboratory at University Putra Malaysia for analysis. Honey stomachs (Figure 1) were obtained through aseptic excisions of the A. dorsata worker bees abdomen in a laminar flow cabinet (Esco, Hatboro, USA).
Figure 1.
The full honey stomach (b) of Apis dorsata (a), including nectar from flowers. The honey stomach is separated from the rest of the digestive tract at the proventriculus (c).
Culture-dependent method
Honey stomach solution (10%, W/V) was prepared in a sterile 0.1% peptone saline solution supplemented with Tween 80 and lactobacilli were isolated from honey stomach on MRS (Deman Rogosa Sharp) agar (Oxoid, Basingstoke, UK). The isolates were incubated for 3–4 days at 37 °C under anaerobic conditions, using Anaerobic Jars with Anaerocult (Merck, Darmstadt, Germany) (Tajabadi et al., 2011b). Next, 92 colonies with varying morphologies were chosen and sub-cultured as pure cultures.
Biochemical screening
Gram staining and catalase test (Coeuret et al., 2003) were conducted as an initial screening of Lactobacillus. Gram-positive, catalase-negative, and bacilli form isolates were chosen. The isolates were maintained as stock cultures at −20 °C in MRS broth (Merck, Darmstadt, Germany) supplemented with 15% glycerol (Merck, Darmstadt, Germany) for further analysis.
DNA extraction
DNA was extracted according to DNA extraction kit protocol (QIAGene, Hilden, Germaney) with some modifications, as described by Ward et al. (Ward et al., 1994). Briefly, all isolated cell were harvested in a microcentrifuge tube, after centrifuging for 10 min at 5000 g. Bacterial pellet was re-suspended in 180 μL enzymatic lysis buffer (20 mM Tris-Cl’ pH = 8.0 -Sigma, Aldrich, USA-, 2 mM sodium salt of ethylenediaminetetraacetic acid (EDTA)-Sigma, Aldrich, USA-, 1.2% Triton X-100 solution-Sigma, Aldrich, USA-, Lysozyme 20 mg/ml -QIAGene, Hilden, Germaney) and incubated for at least 30 min at 37 °C. Twenty five μL proteinase K and 200 μL Buffer AL (QIAGene, Hilden, Germaney) were added into tube and homogenized by vortexing and incubated at 56 °C for 30 min. A volume of 200 μL of ethanol (96%) (HmBg Chemicals, Germany) was added to the sample, and mixed thoroughly by vortexing. The sample was loaded into the DNeasy Mini spin column (QIAGene, Hilden, Germaney) placed in a 2 mL collection tube and centrifuged at 6000 g for 1 min. The DNeasy Mini spin column was transferred in a new 2 mL collection tube, 500 μL Buffer AW1 were added and the resulting solution was centrifuged for 1 min at 6000 g. Next, the DNeasy Mini spin column was placed in a new 2 mL collection tube, 500 μL of buffer AW2 (QIAGene, Hilden, Germaney) were added and the solution was again centrifuged for 3 min at 20,000 g. Next, the DNeasy Mini spin column was placed in a clean 2 mL microcentrifuge tube, and 200 μL of buffer AE were transferred directly to the DNeasy membrane. The sample was incubated at room temperature for 1 min, and then centrifuged for 1 min at 6000 g to elute and stored at −20 °C for further analysis. The degree of DNA purity was determined spectrophotometrically, using 260-nm/280-nm absorbance ratios (Gene Quant pro RNA/DNA Calculator).
PCR reaction and program
In the present study, one set of bacterial universal primers 27F (5’-AGAGTTTGATCCTGG CTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) was used (Lane, 1991). Amplification reactions were performed in a total volume of 25 μL containing 1X Taq Master Mix (New England Biolabs Inc, Ipswich, UK), 1.5 mM MgCl2 (New England Biolabs Inc, Ipswich, UK), 0.25 mM forward primer, 0.25 mM reverse primer, and 0.4 ng of genomic DNA. PCR (Eppendorf, Homburg, Germany) temperature cycling conditions were as follows: an initial heating of 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 55 s, extension at 72 °C for 1 min, and followed by a 10 min final incubation at 72 °C. Gradient temperature PCR was employed to find the optimal annealing temperature for each template-primer pair combination. The PCR products were examined with Gel electrophoresis (Gel Electrophoresis Systems, Major Science, Taiwan). The amplified 16S rDNA amplicons were purified with QIAquick PCR purification kit (QIAGEN, Hilden, Germany), following the manufacturer’s recommendations and homogeneity was then checked through electrophoresis on a 1% w/v agarose gel (First BASE, Kuala Lumpur, Malaysia). The purified products were sequenced by First BASE Laboratories, Malaysia using 27F and 1492R primers. Sequences identification were investigated with GenBank (National Centre for Bio-technology Information, Rockville Pike, Bethesda, MD), employing the advanced BLAST likeness seek option (attainable at http://www.ncbi.nlm.nih.gov/). Phylogenetic analyses were completed, employing the computer software of the neighbor joining method (http://www.megasoftware.net/mega4). The neighbor-joining tree was bootstrapped 1000 times and used from MEGA (4) package (Tamura et al., 2007). All bacterial 16S rDNA gene sequences were entrusted in GenBank using the accession numbers HM027639, HM027640, and HM027642 to HM027644.
Reference sequences used in phylogenetic analysis
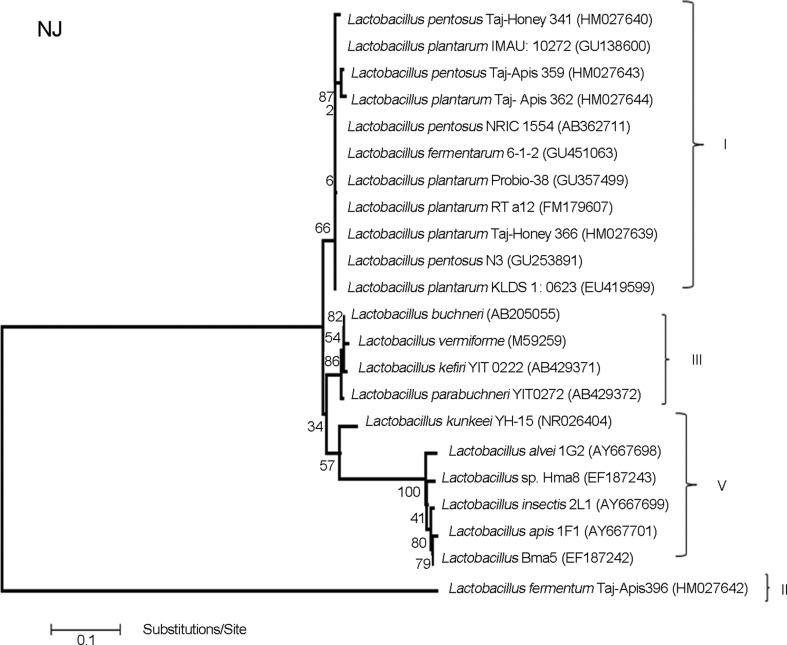
The following bacterial 16S rDNA gene sequences were tested as outgroups in phylogenetic analysis: L. plantarum IMAU 10272 (GU138600), L. pentosus NRIC 1554 (AB362711), L. fermentarum 6–1-2 (GU451063), L. plantarum Probio-38 (GU357499), L. plantarum RT a12 (FM179607), L. pentosus N3 (GU253891), L. plantarum KLDS 1: 0623 (EU419599) (cluster I in Figure 2), L. apis (AY667701), L. alvei (AY667698), L. insectis (AY667699), Lactobacillus sp.Bma5 (EF187242), L. kunkeei YH-15 (NR026404), and Lactobacillus sp. Hma8 (EF187243) (cluster V in Figure 2), L. parabuchneri (AB429372), L. kefiri (AB429371), L. buchneri (AB205055), and L. vermiforme (M59295) (cluster III in Figure 2).
Figure 2.
Phylogenetic analysis of Lactobacillus bacteria in honey stomach of Terengganu. Phylogenetic tree based on a distance matrix analysis of 1,629 positions in the 16S rDNA gene. The phylogenetic tree was constructed by ClustalW using the neighbour-joining method within the MEGA (4) package (Tamura et al., 2007). Closely related type and reference strains are shown in parentheses together with accession numbers from GenBank. Bootstrap values based on 1000 re-samplings display the significance of the interior nodes, and are shown at branch points. Cluster I, L. plantarum and L. pentosus group; cluster II, L. fermentum group; cluster III, Lactobacillus as out group and cluster V, Lactobacillus as out group.
Results
The total 49 sequences isolated from honey stomach of honeybee A. dorsata in Terengganu state showed Gram-positive bacilli and catalase-negative, identified as lacto-bacilli and displayed a similarity of ≥ 99% to type strains deposited in the public GenBank database (http://www.ncbi.nlm.nih.gov/) (Figure 2). Phylogenetic analysis demonstrated that the honey stomach of the Asiatic Giant honeybee A. dorsata was comprised of 5 different phylotypes of Lactobacillus, and 2 of them (Taj-Honey366 and Taj-Apis362) showed 99.74% and 100% similarity with the more closely related strain L. plantarum (clusters I in Figure 2 and Table 1). Moreover, 2 phylotypes, Taj-Honey341 and Taj-Apis359, with 99.5% and 99.42% similarity, were more closely related to L. pentosus (cluster I in Figure 2 and Table 1). The other phylotype, Taj-Apis396 with 99% similarity, was distant but more closely related to L. fermentum (cluster II in Figure 2 and Table 1).
Table 1.
Bacterial strains phylotypes isolated from honey stomach of honeybee Apis dorsata from the forest area of Melaleuca, in Terengganu, Malaysia.
| Isolatesa | Sequence lengths and number of identicalb | Most closely related type strainc | Sequence lengths and similarityd | Accession numberse |
|---|---|---|---|---|
| Taj-Honey366 | (1-1090) (17) | L. plantarum IMAU: 10272(GU138600) | 1106 (99.74) | HM027639 |
| Taj-Honey341 | (1-660) (11) | L. pentosus N3 (GU253891) | 664 (99.5) | HM027640 |
| Taj-Apis396 | (1-938) (1) | L. fermentum (GU451063) | 952 (99) | HM027642 |
| Taj-Apis359 | (1-875) (12) | L. pentosus N3 (GU253891) | 877 (99.42) | HM027643 |
| Taj-Apis362 | (1-812) (8) | L. plantarum KLDS 1.0623(EU419599) | 816 (100) | HM027644 |
The identity of 16S rDNA gene sequences were generated from isolates.
The number of identical sequences found are shown in parentheses and the sequence lengths are shown in brackets.
GenBank references accession numbers are shown in parentheses; taxonomic connection was established by comparing the sequence with those published by the National Center for Biotechnology Information(NCBI).
The similarity to the closest type strain sequence is exhibited as a percentage inside parentheses.
GenBank accession numbers for this study are shown in the last column.
The results of the present study (Figure 2) showed that the lactobacilli present in the honey stomach of Melaleuca secondary forest area is dominated by the Taj-Honey366 phylotype (34.69%), which is most closely related to L. plantarum. Other four dominant lactobacilli in the honey stomach were Taj-Apis359 (24.49%), Taj-Honey341 (22.45%), Taj-Apis362 (16.33%), and Taj-Apis396 (2.04%). The 16S rDNA gene sequences of the new isolates representing different groups and possible new Lactobacillus strains were deposited in GenBank (NCBI) with accession numbers HM027639, HM027640, and HM027642 to HM027644.
Discussion
Lactic acid bacteria are remarkably abundant in honey stomach of honey bees and mainly belong to the genera Lactobacillus, Bifidobacterium, and Enterococcus (Tajabadi et al., 2011a, 2011b).
The genus Lactobacillus, an important group from the LAB, is usually found as commensal bacteria and is commonly used as probiotics in humans and animals (Ouwehand et al., 2002). Lactobacillus originated from the honey stomach of honeybee may be selected and used for food preservation, food enrichment, and food probiotic supplement for human consumption (unpublished data). Historically, most assays to isolate and identify these numerous microorganisms employ culture-based approaches (Gilliam, 1997). More recent assays have applied culture independent 16S rDNA gene sequences analyses to study this community based on single-strand conformation polymorphism. In the present study, classical cultivation procedures has been linked with the 16S rDNA sequencing granted data to describe the bacterial diversity and phylogenetic relationships of Lactobacillus present in the honey stomach of A. dorsata. This study set out with the aim of assessing dominant lactobacilli in the honey stomach of the most popular Malaysian honeybee of A. dorsata in a new complex dynamic ecosystem. In this study, phylogenetic analysis showed that Lactobacillus included five different phylotypes. Dominant strains of lactobacilli (Taj-Honey366 and Taj-Apis362), totalizing 51.02%, were most closely related to the previously described species L. plantarum. This species generally are recognized as safe (GRAS) and several strains of L. plantarum have been isolated from different ecological niches, such as cereal products, fruits, meat, fish, vegetables, and milk. Lactobacillus plantarum has been used as a starter culture in various food fermentation processes, contributing to improve food quality and sensory properties, like flavor, consistency, and texture. Due to the production of lactic acid and other antimicrobial compounds, L. plantarum also might contribute for the safety of the final products (Van Hoorde et al., 2008). The next most dominant lactobacilli in the honey stomach (Taj-Honey341 and Taj-Apis359) were associated with L. pentosus. The species L. pentosus and L. plantarum are genotypically closely related and show highly similar phenotypes. Lactobacillus pentosus isolated from plants has proven to produce the highest bacteriocin titers in environmental conditions (Delgado et al., 2005), which suggest its technological interest as a starter culture. Moreover, the rest phylotype, Taj-Apis396 was distant but most closely related to the L. fermentum. Lactobacillus fermentum is commonly found in fermenting animal and plant material. It has long been used in food processing (LeBlanc et al., 2008) and is well-characterized as a probiotic strain with efficacy in the prevention and treatment of infections in the human being (Gardiner et al., 2002).
Earlier studies established that the typical honeybee gut microorganisms belonging to the genus Lactobacillus, Bacillus, and Bifidobacterium (Rada et al., 1997). Recent studies on the detection and identification of LAB have identified new phylotypes of Lactobacillus in the honey stomach of European honeybee A. mellifera, bumblebee, and A. dorsata (Olofsson and Vasquez, 2008; Vásquez et al., 2009; Tajabadi et al., 2011b). However, the current experiment was the first report highlighting the presence of L. plantarum, L. pentosus, and L. fermentum in the honey stomach of Malaysian honeybee A. dorsata. Olofsson and Vasquez (2008) found different varieties of lactobacilli profiles (L. kunkeei, L. buchneri, and L. acidophilus) in honey stomach of honeybee A. mellifera. In a previous study, we observed different lactobacilli profiles (L. kunkeei, L. vermiform, and Lactobacillus sp) in the honey stomach of honeybees A. dorsata in the highlands of Kedah state, in the north of Malaysia. The result of our previous work in the Kedah state of Malaysia, on the presents of LAB in the honey stomachs of honeybees, in concordance to the studies in the USA and Sweden, those reported that the numbers and members of the LAB microbiota detectable in the honey stomach were seasonal and dependent to the source and amount of nectar (Tajabadi et al., 2011b; Vásquez et al., 2009). Moreover, the presence of other bacterial genera within the honeybees and honeybees species were probably other factors to influence upon composition of LAB in the honey stomach.
Conclusion
In conclusion, our study emphasized on the presence of three promising and novel strains of L. plantarum, L. pentosus, and L. fermentum isolated from the honey stomach of honeybee A. dorsata from the forest area of Melaleuca, in Terengganu, Malaysia. These strains present in honey and resistant to its antimicrobial character could be considered as good candidates for screening of potential probiotics from honeybee and also as candidates as natural food preservatives for their possible use in the food industry. However, studies on the characterization of these novel strains of lactobacilli are recommended.
Acknowledgments
This study had funding support from the Food Bio-technology Research Laboratory, Faculty of Food Sciences and Technology, Universiti Putra Malaysia. The authors would like to thank Dr. A. Javanmard, Mr. M.M. Saberioon and all the honey hunters in Terengganu state of Malaysia for providing honeybee samples.
References
- Caplice E, Fitzgerald G. Food fermentations: role of micro-organisms in food production and preservation. Int J Food Microbiol. 1999;50:131–149. doi: 10.1016/s0168-1605(99)00082-3. [DOI] [PubMed] [Google Scholar]
- Coeuret V, Dubernet S, Bernardeau M, Guegun M, Vernouxj P. Isolation, characterization and identification of lacto-bacilli focusing mainly on cheeses and other dairy products. Lait. 2003;83:269–306. [Google Scholar]
- Delgado A, Brito D, Peres C, Noé-Arroyo F, Garrido-Fernández A. Bacteriocin production by Lactobacillus pentosus B96 can be expressed as a function of temperature and NaCl concentration. Food Microbiol. 2005;22:521–528. [Google Scholar]
- Dsmz. Bacterial nomenclature up to date. 2008 ( http://www.dsmz.de/microorganisms/bacterial_nomenclature)
- Evans J, Lopez D. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae) J Econ Entomol. 2004;97:752–756. doi: 10.1603/0022-0493(2004)097[0752:bpiair]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Forsgren E, Olofsson T, Vásquez A, Fries I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie. 2010;41:99–108. [Google Scholar]
- Fujisawa T, Mitsuoka T. Homofermentative Lactobacillus species predominantly isolated from canine feces. J Vet Med Sci. 1996;58:591–593. doi: 10.1292/jvms.58.591. [DOI] [PubMed] [Google Scholar]
- Gardiner GE, Heinemann C, Bruce AW, Beuerman D, Reid G. Persistence of Lactobacillus fermentum RC-14 and Lactobacillus rhamnosus GR-1 but not Lactobacillus rhamnosus GG in the human vagina as demonstrated by randomly amplified polymorphic DNA. Clin Vaccine Immunol. 2002;9:92–96. doi: 10.1128/CDLI.9.1.92-96.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliam M. Identification and roles of non-pathogenic microflora associated with honey bees. FEMS Microbiol Lett. 1997;155:1–10. [Google Scholar]
- Kandler O, Weiss N, Sneath P, Mair N, Sharpe M, Holt J. Bergey’s manual of systematic bacteriology. Bergey’s manual of systematic bacteriology. 1986:2. [Google Scholar]
- Lane D. In: 16S/23S rRNA sequencing Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. J Wiley and Sons; Chichester: 1991. pp. 115–175. [Google Scholar]
- LeBlanc J, Ledue-Clier F, Bensaada M, de Giori G, Guerekobaya T, Sesma F, Juillard V, Rabot S, Piard J. Ability of Lactobacillus fermentum to overcome host α-galactosidase deficiency, as evidenced by reduction of hydrogen excretion in rats consuming soya α-galacto-oligosaccharides. BMC Microbiol. 2008;8:22. doi: 10.1186/1471-2180-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Kime R. The use of honey for clarifying apple juice. J Apicult Res. 1984;23:45–49. [Google Scholar]
- Ludwig W, Schleifer K. Phylogeny of bacteria beyond the 16S rRNA standard. ASM News. 1999;65:752–757. [Google Scholar]
- Olofsson T, Vasquez A. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr Microbiol. 2008;57:356–363. doi: 10.1007/s00284-008-9202-0. [DOI] [PubMed] [Google Scholar]
- Ouwehand A, Salminen S, Isolauri E. Probiotics: an overview of beneficial effects. Antonie van Leeuwenhoek. 2002;82:279–289. [PubMed] [Google Scholar]
- Rachman C, Kabadjova P, Prevost H, Dousset X. Identification of Lactobacillus alimentarius and Lactobacillus farciminis with 16S–23S rDNA intergenic spacer region polymorphism and PCR amplification using species specific oligonucleotide. J Appl Microbiol. 2003;95:1207–1216. doi: 10.1046/j.1365-2672.2003.02117.x. [DOI] [PubMed] [Google Scholar]
- Rada V, Machova M, Huk J, Marounek M, Duskova D. Microflora in the honeybee digestive tract: counts, characteristics and sensitivity to veterinary drugs. Apidologie. 1997;28:357–365. [Google Scholar]
- Rhee SJ, Lee JE, Lee CH. Importance of lactic acid bacteria in Asian fermented foods. Microb Cell Fact. 2011;10:1–13. doi: 10.1186/1475-2859-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeir J, de Vrese M. Probiotics, prebiotics, and synbiotics-approaching a definition. Am J Clin Nutr. 2001;73:361–364. doi: 10.1093/ajcn/73.2.361s. [DOI] [PubMed] [Google Scholar]
- Settanni L, Van Sinderen D, Rossi J, Corsetti A. Rapid differentiation and in situ detection of 16 sourdough Lactobacillus species by multiplex PCR. Appl Environ Microbiol. 2005;71:3049–3059. doi: 10.1128/AEM.71.6.3049-3059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell-Castro R, Godon JJ, Delgenès JP, Dabert P. Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol Ecol. 2005;52:229–242. doi: 10.1016/j.femsec.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Tajabadi N, Mardan M, Abdul Manap MY, Shuhaimi M. Isolation and Identification of Enterococcus sp. from stomach of honeybee based on biochemical and 16S rRNA sequencing analysis. Int J ProbioPerbio. 2011a;6:95–100. [Google Scholar]
- Tajabadi N, Mardan M, Abdul Manap MY, Shuhaimi M, Meimandipour A, Nateghi L. Detection and identification of Lactobacillus bacteria found in the honey stomach of the giant honeybee Apis dorsata. Apidologie. 2011b;42:642–649. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tannock G. A special fondness for lactobacilli. Appl Environ Microbiol. 2004;70:3189–3194. doi: 10.1128/AEM.70.6.3189-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoorde K, Verstraete T, Vandamme P, Huys G. Diversity of lactic acid bacteria in two Flemish artisan raw milk Gouda-type cheeses. Food Microbiol. 2008;25:929–935. doi: 10.1016/j.fm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Vásquez A, Olofsson T, Sammataro D, Hartfelder K. A scientific note on the lactic acid bacterial flora in honeybees in the USA-A comparison with bees from Sweden. Apidologie. 2009;40:417–417. [Google Scholar]
- Wallbanks S, Martinez-Murcia A, Fryer J, Phillips B, Collins M. 16S rRNA sequence determination for members of the genus Carnobacterium and related lactic acid bacteria and description of Vagococcus salmoninarum sp. nov. Int J Sys Evol Microbiol. 1990;40:224–230. doi: 10.1099/00207713-40-3-224. [DOI] [PubMed] [Google Scholar]
- Ward L, Brown J, Davey G. Application of the ligase chain reaction to the detection of nisinA and nisinZ genes in Lactococcus lactis ssp. lactis. FEMS Microbiol Lett. 1994;117:29–33. doi: 10.1111/j.1574-6968.1994.tb06738.x. [DOI] [PubMed] [Google Scholar]
- Yoshiyama M, Kimura K. Bacteria in the gut of Japanese honeybee, Apis cerana japonica, and their antagonistic effect against Paenibacillus larvae, the causal agent of American foulbrood. J Invert Pathol. 2009;102:91–96. doi: 10.1016/j.jip.2009.07.005. [DOI] [PubMed] [Google Scholar]