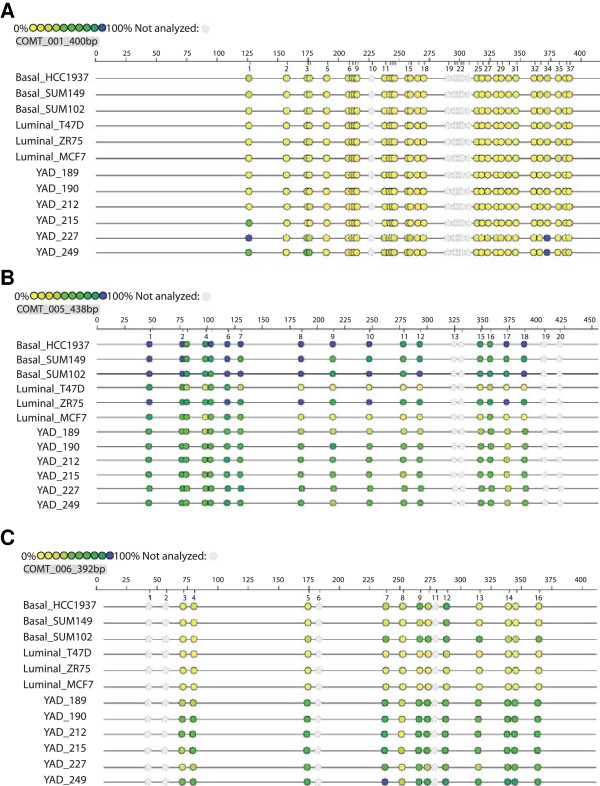
Figure 3.
Epigrams of amplicons illustrate variable methylation by sample type. The Sequenom EpiTYPER MassARRAY platform was used to calculate percent methylation for CpG dinucleotides within each of the three amplicons shown. The epigram schematic illustrates percent methylation for each consecutive CpG (colored circle) on a continuum from yellow (0%) to navy blue (100%). Shaded circles represent CpGs that could not be quantified because they fell outside the Mass Dalton allowable detection window of the EpiTYPER software. Twelve samples per amplicon are shown to illustrate differential methylation: Rows 1–6 represent two groups of biologically distinct breast cancer cell lines (three basal-like and three luminal breast cancer cell lines respectively). Rows 7–12 are from six different healthy saliva samples. A. Amplicon COMT_001 is located in the CpG island 5’ and including exon 1 (Figure 1) and is relatively hypomethylated, with CpGs 1,3,4, and 34 showing the most heterogeneity across sample type. B. Amplicon COMT_005, located in the CpG island within the predicted promoter (P*) for S-COMT, is relatively hypermethylated for CpGs 1–7 for all samples, and most differentially methylated between CpGs 8–18 as illustrated by breast cell line samples T47D & MCF7, & saliva sample YAD189. C. Amplicon COMT_006, is located in Exon 3 (Figure 1), and like COMT_005, is homogeneously methylated at the beginning of the amplicon (CpGs 1–6), with differential methylation occurring between CpGs 7–16, as evidenced by relative hypermethylation of these loci in the basal versus the luminal cell lines.