Fig. 3.
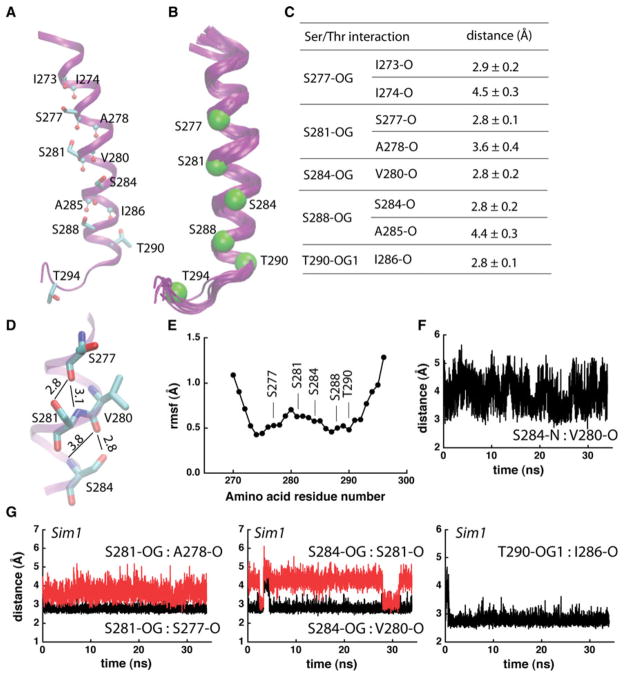
Intrahelical hydrogen bonding in Sim1. a Each of the Ser amino acids from the TM helical part of peptide 1 (Table 1) engages in at least 1 intrahelical hydrogen bond. Ser/Thr groups are shown as bonds, and Leu backbone groups that hydrogen bond with Ser/Thr are shown with atoms as small spheres. b Overlap of 10 snapshots of the last 10 ns of Sim1 taken at every 1 ns. Cα atoms of the Ser/Thr amino acid residues at the end of Sim1 are shown as Van der Waals spheres. c Average distances between Ser/Thr hydroxyl oxygen atoms and their interaction partners. Average and standard deviation values were computed from the last 10 ns of Sim1 (1000 coordinate sets). d Close view of hydrogen bonding in the middle segment of peptide 1. Thin lines with numbers indicate hydrogen bonding and the corresponding distances in Å. The standard deviations for the side chain distances are given in (c); for the backbone S281-N:S277-O, and S284-N:V280-O distances, the standard deviations are ±0.3 Å and ±0.5 Å, respectively. e Cα-rmsf profile of peptide 1, computed from the last 10 ns segment of Sim1. The rmsf profile and the overlap of structural snapshots in (b) would suggest an enhanced flexibility of the Ser/Thr-rich TM helix. f Time series of the S284-N:V280-O distance. g Examples of distances (in Å) between Ser/Thr hydroxyl groups and backbone carbonyl groups monitored during Sim1. For all time series presented here and in Fig. 4, the origin of the time axis corresponds to all harmonic constraints being switched off