Abstract
The search for new tuberculosis treatments continues as we need to find molecules that can act more quickly, be accommodated in multi-drug regimens, and overcome ever increasing levels of drug resistance. Multiple large scale phenotypic high-throughput screens against Mycobacterium tuberculosis (Mtb) have generated dose response data, enabling the generation of machine learning models. These models also incorporated cytotoxicity data and were recently validated with a large external dataset.
A cheminformatics data-fusion approach followed by Bayesian machine learning, Support Vector Machine or Recursive Partitioning model development (based on publicly available Mtb screening data) was used to compare individual datasets and subsequent combined models. A set of 1924 commercially available molecules with promising antitubercular activity (and lack of relative cytotoxicity to Vero cells) were used to evaluate the predictive nature of the models. We demonstrate that combining three datasets incorporating antitubercular and cytotoxicity data in Vero cells from our previous screens results in external validation receiver operator curve (ROC) of 0.83 (Bayesian or RP Forest). Models that do not have the highest five-fold cross validation ROC scores can outperform other models in a test set dependent manner.
We demonstrate with predictions for a recently published set of Mtb leads from GlaxoSmithKline that no single machine learning model may be enough to identify compounds of interest. Dataset fusion represents a further useful strategy for machine learning construction as illustrated with Mtb. Coverage of chemistry and Mtb target spaces may also be limiting factors for the whole-cell screening data generated to date.
Keywords: Bayesian models, Collaborative Drug Discovery Tuberculosis database, Dual-event models, Function class fingerprints, Lead optimization, Mycobacterium tuberculosis, Recursive partitioning, Support vector machine, Tuberculosis
INTRODUCTION
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), infects approximately one third of the world’s population, and 1.7–1.8 million people die from this disease annually 1. Agents active against Mtb are urgently needed to overcome resistance to the available regimen of drugs, shorten a lengthy treatment (that is at a minimum six months in duration), and address drug-drug interactions that may arise during the treatment of TB/HIV co-infections 2, 3. Efforts to leverage sequencing and partial annotation of the Mtb genome 4 and pursue specific small molecule modulators of the function of essential gene products have proven more challenging than expected 5, 6 in part due to a suggested disconnect between inhibition of protein function and a no-growth whole-cell phenotype 7. Thus, a target-agnostic approach has gained favor in recent years, focusing on whole-cell phenotypic highthroughput screens (HTS) of commercial vendor libraries 3, 8–10. This random approach has afforded the clinical-stage SQ109 11 and a diarylquinoline hit that was optimized to afford the drug bedaquiline 12. However, Mtb screening hit rates tend to be in the low single digits, if not below 1% as seen elsewhere in drug discovery 13.
One can, however, learn from both the active and inactive samples arising from these screens. Leveraging this prior knowledge to produce computational models is an approach we have taken to improve screening efficiency both in terms of cost and relative hit rates. Machine learning and classification methods have been used in TB drug discovery 14, and have enabled rapid virtual screening of compound libraries for novel inhibitors 15, 16. Specifically, Novartis examined the application of Bayesian models, relying on conditional probabilities 17. Our work has built on this early contribution to examine significantly larger screening libraries (individually in excess of 200,000 compounds) utilizing commercially available model construction software with molecular function class fingerprints of maximum diameter 6 (FCFP_6) 18 to model recent tuberculosis screening datasets 19–21. Single- (predicting whole-cell antitubercular activity) and dual-event (predicting both efficacy and lack of model mammalian cell line cytotoxicity where: IC90 < 10 µg/ml or 10 µM and a selectivity index (SI) greater than ten where the SI is calculated from SI = CC50/IC90) have been created 9. The models were demonstrated to be statistically robust 17 and validated retrospectively through enrichment studies (in excess of 10-fold as compared to random HTS) 20. Most significantly, the Bayesian models were harnessed to predict novel actives through experimental validation with hit rates up to ~20%. 22, 23. Most recently we examined 1924 molecules with three dual-event dose response and cytotoxicity models (these are called MLSMR (derived from Molecular Libraries Screening Center Network), TAACF-CB2, and TAACF kinase) 24. The molecules were ranked using the Bayesian score (which scales with the probability of activity) from all three different dual-event models. Then a receiver operator curve (ROC) plot was generated and we found the MLSMR dose response and cytotoxicity model appeared to perform the best at identifying the active compounds (11.8 fold enrichment in the top 1%). The TAACF kinase dose response and cytotoxicity model showed a similar enrichment (11.1 fold) while the TAACF-CB2 dose response and cytotoxicity model consistently performed poorly. These results highlighted the influence of model training set on performance, suggesting the utility of using multiple models as it is not known a priori which model may perform the best. We now evaluate the effect of combination of datasets and use of different machine learning algorithms (Support Vector Machines, Recursive Partitioning (RP) Forests, RP Single Trees and Bayesian) and their impact on model predictions (internal and external validation) using data from the same laboratory (to minimize inter-laboratory variability 25) and the literature. The knowledge gained from these studies will aid in the further development of machine-learning methods with tuberculosis drug discovery.
MATERIALS AND METHODS
CDD Database and SRI Datasets
The development of the CDD TB database (Collaborative Drug Discovery Inc. Burlingame, CA) has been previously described 21. The Tuberculosis Antimicrobial Acquisition and Coordinating Facility (TAACF) and Molecular Libraries Small Molecule Repository (MLSMR) screening datasets 8–10 were collected and uploaded in CDD TB from sdf files and mapped to custom protocols 26. All of these Mtb datasets used in model building are available for free public read-only access and mining upon registration in the CDD database 20, 26–28, making them a valuable molecule resource for researchers along with available contextual data on these samples from other non Mtb assays. These datasets used previously for modeling are also publically available in PubChem 29. The TB: ARRA dataset used as a test set is available in the CDD TB database (Collaborative Drug Discovery, Burlingame, CA) 24, 26.
Building and Validating Dual-Event Machine Learning Models with Novel Bioactivity and Cytotoxicity Data
We have previously described the generation and validation of the Laplacian-corrected Bayesian classifier models developed with cytotoxicity data to create dual-event models 22, 23 using Discovery Studio 3.5 (San Diego, CA) 17, 30–33. These models were developed based on: a. MLSMR dose response and cytotoxicity; b. TAACF-CB2 dose response and cytotoxicity; and c. TAACF kinase dose response and cytotoxicity, where cytotoxicity was determined in Vero cells for each set. All three models were generated using standard protocols with the following molecular descriptors: molecular function class fingerprints of maximum diameter 6 (FCFP_6) 18, AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area were calculated from input sdf files. Models were validated using leave-one-out cross-validation in which each sample was left out one at a time, a model was built using the remaining samples, and that model utilized to predict the left-out sample. Each model was internally validated, ROC plots were generated, and the crossvalidated ROC area under the curve (XV ROC AUC) calculated. All Bayesian models generated were additionally evaluated by leaving out 50% of the data and rebuilding the model 100 times using a custom protocol for validation, to generate the ROC AUC, concordance, specificity and selectivity as described previously 22, 23. The three models were used to score a set of 1924 commercial analogs previously in the ARRA dataset 24. In addition we used the ARRA dataset to create a separate dual-event model. The prediction data were evaluated using a receiver operator characteristic (ROC) plot. In the current study, as well as using the datasets individually, we also combined the three previously generated datasets (MLSMR, TAACF-CB2, TAACF-kinase) and compared Bayesian, SVM and RP Forest and single tree models built with the same molecular descriptors in Discovery Studio. For SVM models we calculated interpretable descriptors in Discovery Studio then used Pipeline Pilot to generate the FCFP_6 descriptors followed by integration with R 34. RP Forest and RP Single Tree models used the standard protocol in Discovery Studio. In the case of RP Forest models 10 trees were created with bagging. Bagging is short for “Bootstrap AGgregation”. For each tree, a bootstrap sample of the original data is taken, and this sample is used to grow the tree. A bootstrap sample is a data set of the same size as the original one, but in which the same data record can be included multiple times. RP Single Trees had a minimum of 10 samples per node and a maximum tree depth of 20. In all cases, 5-fold cross validation (leave out 20% of the database) was used to calculate the ROC for the models generated. In the case of the combined datasets, predictions were evaluated using binary classification as well as the continuous probability score calculated where possible (e.g. Bayesian Score) followed by ROC plot calculation.
Testing Machine Learning Models with Additional Previously Published Data and Assessing Chemistry Space
177 Mtb leads were recently disclosed by GlaxoSmithKline (GSK) 35 and represent a promising set of small molecules for further exploration as potential antitubercular drug candidates. The GSK set was scored with all of the combined models generated in this study. As the 177 compounds can be classed as actives, our goal was to ascertain which models were able to predict the most as actives. In addition, we compared the 177 compounds to the four datasets used in this study (including actives and inactives) as to their relative placement in chemistry space. We generated a Principal Component Analysis (PCA) using Discovery Studio with the interpretable descriptors chosen previously (AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area). The mean closest distance to training set was also calculated for the 177 compounds for each of the five models to provide an idea of similarity of the test set to the training set. These data were calculated from the outputs of each of the Bayesian models. For each test set molecule a score for closest distance to training set was calculated using Discovery Studio. We averaged this number across the 177 molecules. The smaller the value, the closer a compound is to the training set. In the past we had used mean-maximal similarity value which provides a value of the opposite magnitude.
Understanding the Mtb Target Space Using Known Inhibitors
745 compounds with known Mtb targets collated from the literature 36 and available in TB Mobile 37 were utilized to generate a PCA plot with the interpretable descriptors selected previously (AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area) for machine learning. This PCA model represents essentially the published target-chemistry space for Mtb. We also compared 1429 Mtb hits (active and non-toxic only, from the SRI screens where: IC90 < 10 µg/ml or 10 µM and a selectivity index (SI) greater than ten where the SI is calculated from SI = CC50/IC90) to show how they covered the target-chemistry space. In addition the 177 GSK Mtb leads published by GSK recently 35 were also compared to this target-chemistry space using PCA. The overlaps in data sets were qualitatively compared.
RESULTS
Effect of Training Set and Approach on Prediction of ARRA data
Following on from a previous study in which a large external set of 1924 molecules (ARRA) was used to evaluate three Bayesian models by assessing the enrichment in finding active compounds, we calculated ROC AUC values using the Bayesian score for ranking compounds (Table 1) 24. The MLSMR dose response and cytotoxicity model had the best value (0.82) followed by the TAACF kinase dose response and cytotoxicity model (0.74) and these data are in line with the enrichments we observed previously 24 (Table 1). In addition, these values were similar if not identical to the ROC AUC values for leave out 50% × 100 cross validation performed previously 22, 23. This comparison of models stimulated us to explore different machine learning models and combining data sets as well as suggested that leave out cross validation provided similar results to using a single external test set. The TAACF-CB2 models performed poorly as described previously 24.
Table 1.
Bayesian models predicting the ARRA dose response and cytotoxicity data. Where: IC90 < 10 µg/ml (TAACF-CB2) or 10 µM and a selectivity index (SI) greater than ten were the SI is calculated from SI = CC50/IC90. Receiver Operator Curve Statistics were calculated for previously published data 22, 23.
|
Mtb Models
(training set N) |
Bayesian (Leave out 50% × 100 ROC) |
Predicting ‘ARRA dose response and cytotoxicity’ dataset (N = 1924) ROC |
Enrichment observed in top 20 ranked ‘ARRA dose response and cytotoxicity’ dataset molecules (Vero, THP-1 and HepG2 cell data) 24 |
|---|---|---|---|
| MLSMR dose response and cytotoxicity (2273) | 0.82 22 | 0.82 | 10.7 – 11.8 fold |
| TAACF-CB2 dose response and cytotoxicity (1783) | 0.64 23 | 0.54 | Poor – random |
| TAACF Kinase dose response and cytotoxicity (1248) | 0.74 22 | 0.74 | 6.7–11.1 fold |
Comparing SVM, Trees and Bayesian Dual Event Machine Learning Models
Ligand based screening studies traditionally use one or more machine learning approach to build models and predict new compounds, with individual groups having their own preferred methods. Previously we have reported the use of one such approach applied to Mtb, namely, Bayesian models. To insure that our studies of training set effects are more broadly applicable, we now report the examination of SVM, RP Single Tree and RP forest models to compare with Bayesian models. These types of models (Bayesian, SVM, and RP) are the most commonly used of machine learning methods and offer documented differences in terms of their approach and ability to fit the training set data versus offer predictive capability outside of the training set’s chemical space 38. RP models are easily interpretable, while also providing a high degree of predictive accuracy. Single Tree models can be influenced by small changes in the training data resulting in a large change in the tree, and, hence, poorer resulting predictions. An RP forest model resamples the training data randomly multiple times and then grows a tree from each resampled dataset. When making predictions the sample is sent down each tree until it reaches a leaf node then the leaf node probabilities are averaged together to yield a prediction for the forest. SVMs have been widely described in the literature and at their core is the use of a kernel function which converts a scalar product into a higher dimensional space to attempt a linear separation (summarized previously 39). SVMs are generally used for binary data and ranking.
The new machine learning models were generated with all three original datasets (MLSMR, TAACF-CB2, and TAACF kinase; dose-response and cytotoxicity) as well as the more recent ARRA dataset. The Bayesian model statistics were generated by leaving out 50% of the data and rebuilding the model 100 times using a custom protocol for validation to generate the ROC AUC, concordance, specificity and sensitivity as described previously 22, 23, are shown in Supplemental Table 1. Using the FCFP-6 descriptors, we can identify those substructure descriptors consistent with both activity and lack of cytotoxicity, namely alkyl-2-aryloxyacetate and 2,4-disubstituted 1,3,4-oxadiazole (Figure S1), and features of inactives such as 2,5-disubstituted furan, oxepane, tetrasubstituted pyrazole/pyrazolidine, 5-substituted 1,3,4-oxadiazole 2-amide and 2-substituted thiazole/thiazolidine (Figure S2).
For comparison of all the machine learning models we used a slightly less aggressive cross validation (5 fold, e.g. leave out 20%) as this is readily implemented in the machine learning methods. The models provide almost identical ROC AUC results with the leave out 50% × 100 when performed with the datasets (Tables 1 and 2). The RP Forest method used an out-of-bag ROC (in which 20% of the compounds are left out from model building). All four machine learning methods show comparable ROC AUC values across the four datasets using this method of internal validation. The Bayesian method has the best statistics based on the 5-fold cross validation with ROC values slightly higher across all models.
Table 2.
Individual machine learning model cross validation Receiver Operator Curve Statistics. Where: IC90 < 10µg/ml (CB2-TAACF) or 10µM and a selectivity index (SI) greater than ten were the SI is calculated from SI = CC50/IC90.
|
Mtb Models
(training set N) |
RP Forest (Out of bag ROC) |
RP Single Tree (With 5 fold cross validation ROC) |
SVM (with 5 fold cross validation ROC) |
Bayesian (with 5 fold cross validation ROC) |
Bayesian (leave out 50% × 100 ROC) |
|---|---|---|---|---|---|
| MLSMR dose response and cytotoxicity (2273) | 0.78 | 0.77 | 0.80 | 0.83 | 0.82 |
| TAACF-CB2 dose response and cytotoxicity (1783) | 0.57 | 0.57 | 0.58 | 0.60 | 0.64 |
| TAACF Kinase dose response and cytotoxicity (1248) | 0.73 | 0.72 | 0.75 | 0.76 | 0.74 |
| ARRA dose response and cytotoxicity (1924) | 0.82 | 0.80 | 0.83 | 0.86 | 0.81 |
The three original data sets (MLSMR, TAACF-CB2, and TAACF kinase; dose-response and cytotoxicity) were combined to build SVM, RP Forest, RP Single Tree and Bayesian models that were then used to predict the ARRA dataset. The Bayesian model statistics for the combined model were generated by leaving out 50% of the data and rebuilding the model 100 times, using a custom protocol for validation. The ROC AUC, concordance, specificity and sensitivity, described previously 22, 23, are shown in Supplemental Table 1. Using the FCFP-6 descriptors, we can identify those substructure descriptors consistent with both activity and lack of cytotoxicity including 3,5-disubstituted thienopyrimidinone, 1-adamantane and acylthiourea (Figure S3) and features of inactives such as isothiazole/isothiazolidine, benzoisoxazole and pyrazoloquinoline (Figure S4).
The external testing ROC AUC for combined models using the ARRA dataset with Bayesian, RP Forest and RP Single Tree methods ranged from 0.65–0.83 for probability (Trees) or Bayesian scores data (Table 3). The SVM method used did not output a continuous probability in the implementation used and so was excluded from this comparison. While using the predicted classification data for the ARRA dataset for all 4 machine learning methods was more instructive (Table 4). For example the Bayesian method had the worst concordance and specificity but the best sensitivity (92.7%) while the SVM had the best concordance and specificity. The RP Single Tree had the lowest sensitivity (58.5%) (Table 4).
Table 3.
Combined MLSMR, TAACF-CB2 and TAACF Kinase dose response and cytotoxicity dataset models created with RP Forest models (Out of bag testing ROC = 0.71), RP Single Tree (Out of bag testing ROC = 0.74) and Bayesian (5 fold cross validation ROC = 0.75) used to predict the ARRA dose response and cytotoxicity data, reporting Receiver Operator Curve statistics using probability (Trees) or Bayesian scores. Note SVM model did not out put a probability value.
| Mtb Models | ROC AUC |
|---|---|
| RP Forest | 0.83 |
| RP Single Tree | 0.65 |
| Bayesian | 0.83 |
Table 4.
Combined MLSMR, TAACF-CB2 and TAACF Kinase dose response and cytotoxicity dataset models created with SVM (5 fold cross validation ROC = 0.73), RP Forest models (Out of bag testing ROC = 0.71), RP Single Tree (Out of bag testing ROC = 0.74) and Bayesian (5 fold cross validation ROC = 0.75) used to predict the ARRA dose response and cytotoxicity data, reporting contingency table statistics for classification data.
| Mtb Models | Concordance (%) |
Specificity (%) |
Sensitivity (%) |
|---|---|---|---|
| SVM | 76.7 | 77.1 | 67.1 |
| RP Forest | 63.1 | 61.9 | 89.0 |
| RP Single Tree | 69.1 | 69.5 | 58.5 |
| Bayesian | 47.2 | 45.2 | 92.7 |
The Effect of Training Set Selection on Prediction of GSK Data and Assessment of Mtb Chemistry Space
The 177 Mtb leads published by GSK recently 35 were scored with the combined models generated in this study (Supplemental Table 2). As all of the 177 compounds can be classed as actives, our goal was to ascertain which models were able to predict the most as actives. We found the TAACF-CB2 dose response and cytotoxicity models performed best, correctly identifying between 48–67.8% of the compounds (Table 5). The SVM model performed optimally with this test set. It is important to note that out of the 177 GSK compounds only a small number were in the models (MLSMR N = 5, TAACF-CB2 N = 2, TAACF-Kinase N = 3, ARRA N = 4, and combined N = 10).
Table 5.
The number of molecules predicted as active out of 177 GSK 35 lead compounds (%). Mean-closest distance = smaller is more similar to training set. Out of the 177 GSK compounds only a small number were in the models corresponding to MLSMR (N = 5), TAACF=CB2 (N = 2), SRI-Kinase (N = 3), ARRA (N = 4) and combined (N = 10). These were included in the table above for ease of comparison.
| Mtb Models (training set N) | Random Forest |
SVM | Bayesian | Mean–closest distance of training set to test set |
|---|---|---|---|---|
| MLSMR dose response and cytotoxicity (2273) | 17 (9.6) | 12 (6.8) | 66 (37.3) | 0.50 |
| TAACF-CB2 dose response and cytotoxicity (1783) | 97 (54.8) | 120 (67.8) | 85 (48.0) | 0.58 |
| TAACF Kinase dose response and cytotoxicity (1248) | 36 (20.3) | 1 (0.5) | 33 (18.6) | 0.62 |
| ARRA dose response and cytotoxicity (1924) | 7 (3.9) | 0 (0) | 17 (9.6) | 0.59 |
| Combined MLSMR, TAACF-CB2 and TAACF Kinase dose response and cytotoxicity | 34 (19.2) | 23 (13) | 65 (36.7) | 0.46 |
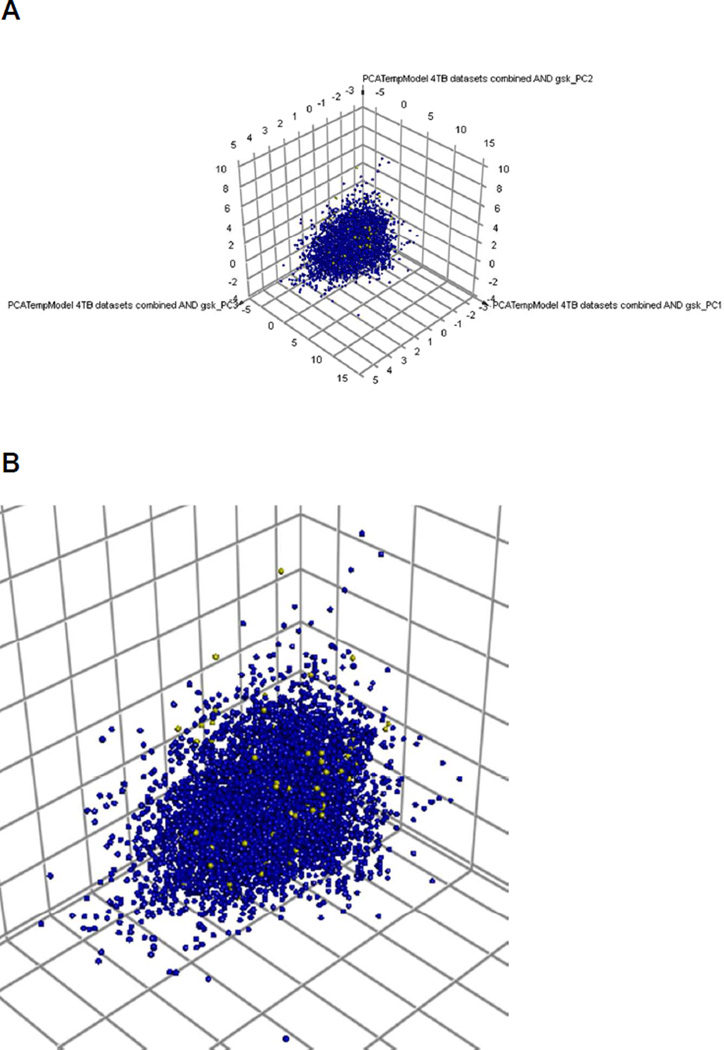
A comparison was made of the 177 compounds to all four datasets used in this study with a Principal Component Analysis. The GSK leads appear distributed within the chemistry space of the >7000 compounds (Figure 1). Next we calculated the mean closest distance to the model training set for each of the 177 compounds to provide an idea of similarity of the test set to the training set. All datasets have roughly similar values but the test set was closest to the combined dataset based on this measure of similarity, while the TAACF-CB2 dose response and cytotoxicity dataset was third closest to the GSK hits. This may suggest such similarity predictors are not a valid measure of model success alone.
Figure 1.
A. Principal Component Analysis of all Mtb datasets (7728 active and inactive compounds) used in this study and overlap of 177 GSK published leads. 3 principal components explain 73% of the variance. B inset to show some of the GSK leads (yellow) widely dispersed and within the chemistry space of the Mtb datasets used for modeling.
Understanding the Mtb target Space Using Known Ligands
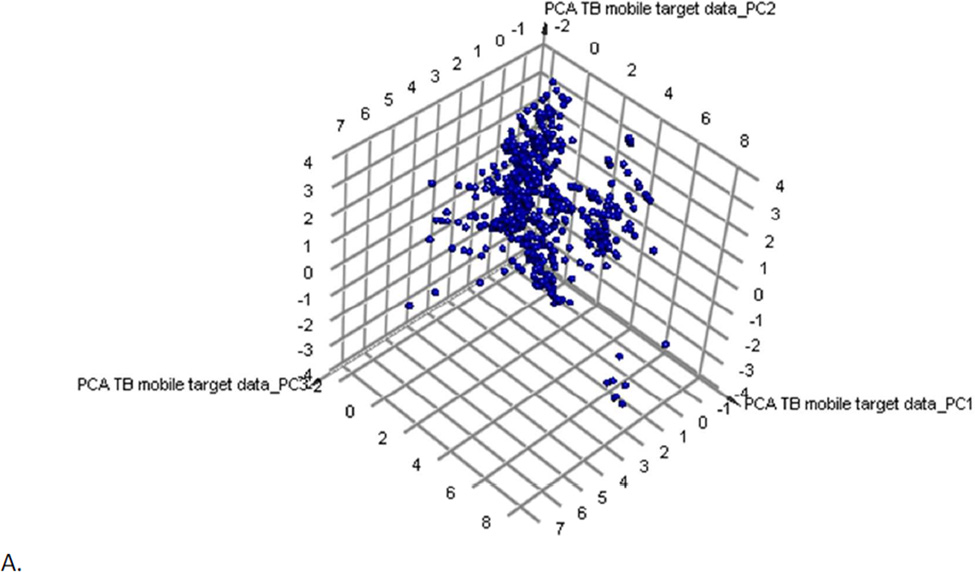
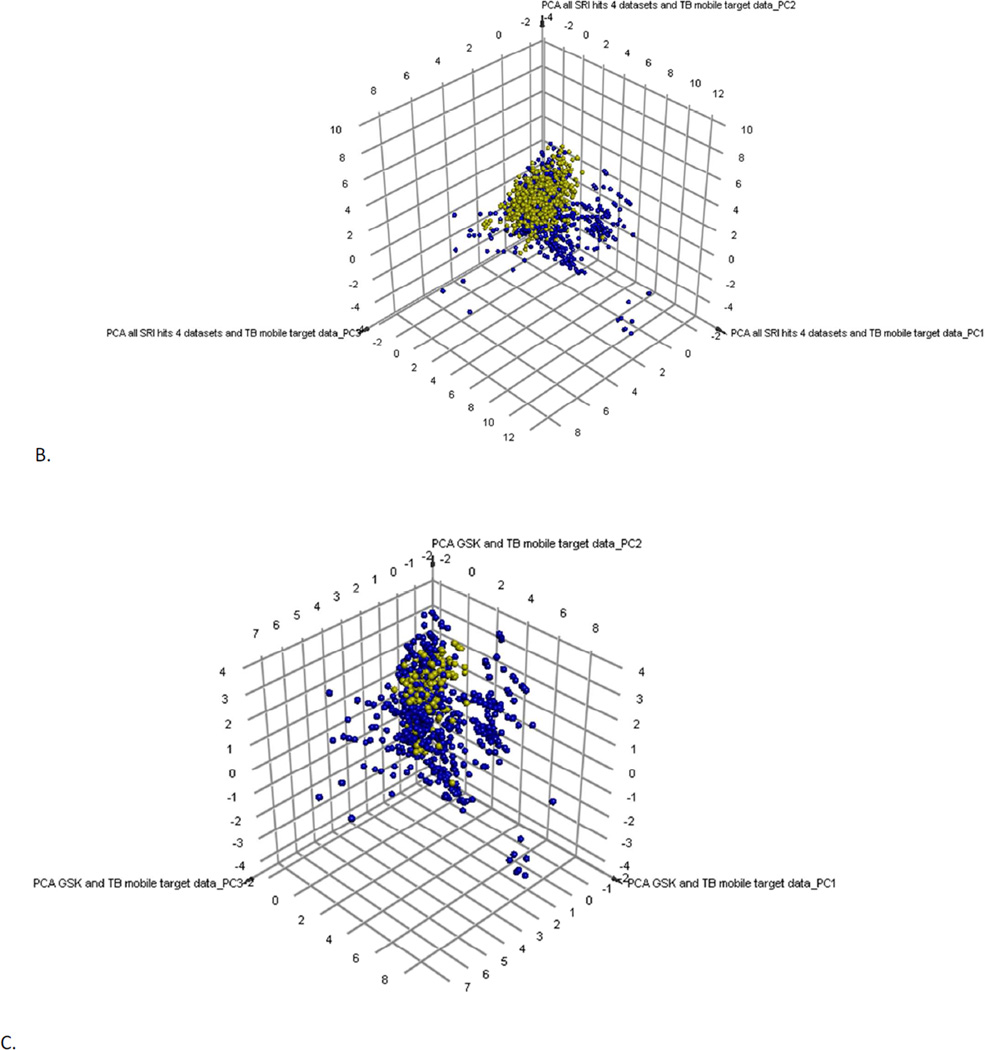
We previously created a collection of molecules with their Mtb target/s from published data 28 collated in the course of a previous study 36. This dataset was made available in the Collaborative Drug Discovery (CDD) database 28 and most recently the TB Mobile app 37. We have recently updated the content such that we have 745 small molecules. Following PCA these compounds can give us an approximation of target chemistry space covered in the literature for known antituberculars (Figure 2). When we overlap the 1429 SRI (active and non-cytotoxic compounds) obtained from the 4 different datasets (based on the previously described methods) they overlap approximately half of the compounds with target data (Figure 2B). The 177 GSK hits overlap partially the same area as the SRI hits, but they cover less space in the plot. The GSK hits were also clustered with the 745 compounds with known Mtb targets as a method to infer their potential targets (Supplemental Table 3). Clustering used the MDL fingerprints and created 100 clusters. Examples of compounds clustering near molecules with known targets in Mtb are shown in Figure S5. These include compounds clustering near known QcrB inhibitors (Figure S5A), PanC inhibitors (Figure S5B), Alr or IlvG (Figure S5C), MmpL3 (Figure S5D), Alr (Figure S5E) and InhA (Figure S5F).
Figure 2.
Clustering and PCA of TB Mobile data. A. Examination of 745 TB Mobile molecules with interpretable descriptors results in a PCA with 3 PCs, which explain 88% variability. Outlier compounds represent macrocycles (bottom right) and long lipid-like molecules (bottom left). B. 1429 SRI hits from four datasets (active and non-toxic only, from the SRI screens where: IC90 < 10 µg/ml or 10 µM and a selectivity index (SI) greater than ten where the SI is calculated from SI = CC50/IC90) and 745 TB Mobile compounds results in a PCA with 3 PCs explaining 83% variability; SRI compounds are clustered (yellow). C. Examination of 177 GSK leads (yellow) and the TB Mobile compounds results in a PCA with 3 PCs, which explain 88 % of variance.
DISCUSSION
There is a resurgence in whole cell HTS for Mtb and this has resulted in low hit rates 35, 40–42. Utilizing past screening data with machine learning methods could improve the efficiency of such screens. Our prior machine learning studies have demonstrated that single and dual-event Bayesian machine learning models based on public data can enrich hit discovery using retrospective and prospective testing 22, 23. While we have focused on Bayesian machine learning due to their processing speed and ease of use, many other algorithms exist that can be used for machine learning. SVM 43–52 and Random Forests 53–55 like Bayesian classification methods 56–60 have also been used extensively for drug discovery and ADME/Tox models 31, 57, 61, 62. For example, extensive evaluations of different machine learning methods and descriptors have been performed by Broccatelli et al. 63 using SVM, Random Forest, Partial Least Squares, Linear Discriminant Analysis, Random Forests (RF) and Genetic Algorithm-kNN models with MOE, MACCS, CDK, Dragon descriptors and 545 literature compounds with the ion channel hERG activity. The best models were RF MOE2D, RF-MACCS and PLSD-VS+ with consensus accuracy 90%, specificity 93% and sensitivity 89%. A set of 7617 compounds with genotoxicity (Ames) data were used to compare five machine learning methods (SVM, kNN, Naïve Bayes, Artificial Neural networks and C4.5 decision trees) each using five fingerprint descriptor methods (PubChem, E-state, MACCS, CDK fingerprints and substructure fingerprints) 64. Using a test set of 831 diverse molecules, the accuracy ranged from 90–98% with three combinations of descriptors and algorithms proving equally accurate (PubChem-kNN, MAACS-kNN and PubChem SVM). Although we have analyzed the Mtb literature extensively 65, 66 we are not aware of similar exhaustive analyses of machine learning methods used to prospectively predict whole cell Mtb activity. Predominantly the focus has been retrospective or leave out testing 67,68
Frequently, we have seen multiple Bayesian models perform differently with varying datasets 19–24 and with the current test set we see a wide range in the ROC values for the ARRA dataset of 1924 molecules, with ROC AUC values of 0.54 – 0.82 (Table 1, not previously reported). Interestingly, combining the datasets only slightly improves the Bayesian model ROC value to 0.83 (Table 1 versus Table 3). However, this model also has the lowest concordance when compared to the other methods at binary classification of the 1924 compounds (Table 4). Using an external dataset of 177 recently published Mtb leads from GSK 35 we found a wide variability between models and datasets in identifying leads from this set (Table 4). It should also be noted that all these molecules can be classed as actives while only a small number of compounds overlapped between the training and test sets. The best models at evaluating this GSK test set, identifying approximately 48–68% of the actives, were the TAACF-CB2 dose response and cytotoxicity RP Forest, SVM, and Bayesian models. These highlight the value of using such models to select compounds for testing without extensive HTS. We had previously used the Bayesian model successfully to screen a larger set of 13,533 GSK compounds found to have antimalarial activity 69. We had scored these molecules 70, which enabled us to identify several with potent antitubercular activity upon empirical testing 23. Yet, this present work also suggests using the ROC value for 5-fold validation alone is not likely to be a single reliable measure (or predictor) of the utility of a model as this TAACF-CB2 dose response and cytotoxicity model also had the lowest ROC scores (below 0.6, Table 2). Conversely, we have also shown that the similarity of molecules in the test and training sets is also not a reliable measure of likely correct predictions as the TAACF-CB2 training set was not the closest to the test set of the GSK leads (Table 4). This result may also suggest the need for a deeper analysis of FCFP_6 descriptors between training and test sets, or more simply a further investigation as to which molecular substructures are important for Mtb activity (that are present in the training and test set molecules). Overlap of certain molecular features between datasets may be a better predictor of the ROC value and model performance (Figure S1–4) and this hypothesis remains to be tested. Ultimately in comparing predictions across datasets one also should consider experimental variability in Mtb screening 25, so it is at least reassuring that models from one laboratory can be used to predict data from another to a reasonable degree. Of course we have relied in this study on the ROC metric (Tables 1–3) and contingency table statistics (Table 4) as measures for comparing models. This may not be enough. Future studies could explore whether other measures commonly used for assessment of virtual screening provide more insight into why there are model and dataset dependencies (e.g. concentrated ROC (CROC), Boltzmann-enhanced discrimination of ROC (BEDROC), Guner Henry Score etc.) 71–74 and whether consensus scoring could overcome these.
This study continues our efforts to build and validate machine learning models for Mtb. 19–24 It extends recent externally validated dual-event models to consider the fusion of datasets as a method to increase coverage of chemistry space and simplify the number of models required. Although, it should be noted that the MLSMR, TAACF-CB2 and TAACF kinase datasets have a fair degree of overlap, and the ARRA dataset overlaps with some of these 24, which may explain why the ROC AUC values for this dataset vary from 0.54 – 0.83 when looking at individual models (Table 1) and there is not a great deal of improvement when datasets are combined. There is also some variation in ROC AUC values across machine learning models when the datasets are combined (Table 3) and across contingency table statistics (Table 4).
Our PCA in this study using molecules with annotated targets (covering over 70 to date with identified inhibitors 37) suggests the hits from SRI and GSK overlap and are only exploring a fraction of the Mtb chemistry target space. So this might indicate that any machine learning models derived from such HTS data are only going to be useful for predictions in a relatively small segment of Mtb chemistry target space. Conversely, this type of analysis may also be useful for predicting potential targets for the training set actives. The opportunity also exists to extend our initial approach based on molecule similarity 37 to one predicated on multiple physicochemical descriptors. The potential targets for some of the 177 GSK compounds are suggested based on clustering with compounds with known annotated Mtb targets which could be useful for further future experimental verification. Similarly one could pursue this approach with the active subset of compounds in the ARRA or other datasets. Our approach in this study using machine learning models to predict compounds with activity could also be combined with inhibitors of known targets and clustering to suggest their potential targets in a single workflow. Such a process may lead to more rapid target identification efforts. Verification of such predictions is however time consuming and costly and whole cell phenotypic screening will also identify compounds that act through more than one mechanism.
In conclusion, the choice of Bayesian models would appear to be acceptable for predicting whole-cell antitubercular efficacy under the current conditions when compared to SVM and RP approaches. Each of the methods has their strengths and weaknesses and it would appear that no one method stands out as best for Mtb active prediction. Others have previously shown SVM and Random Forest approaches to outperform Bayesian models in different areas 64. Additional researchers have used ensembles of models rather than rely on a single model 75. To date none of these ensemble machine learning approaches had been tested with Mtb datasets. A major advantage of dataset fusion is that a single model can be created that covers the sum chemical space of individual models and may be more likely to be used rather than multiple individual smaller models. This is distinct from the fusion of predictions and consensus scoring with individual machine learning or similarity methods 76. Future efforts may explore using other machine learning methods, e.g. k–Nearest Neighbors 77, K-Partial Least Squares 78, Self Organizing Maps and Kohonen maps 79 for Mtb model building with this combined dataset. In addition, efforts to make Mtb models more readily available may also be evaluated using free or open source resources like Bioclipse 80–82, Chembench 83 and others 84, 85. This would then make the models globally accessible 86 and perhaps increase the speed and efficiency of screening efforts in vitro.
Supplementary Material
Acknowledgment
S.E. acknowledges colleagues at CDD. Accelrys are kindly acknowledged for providing Discovery Studio and Dr. Katalin Nadassy for her support. The Bayesian models created in Discovery Studio are available from the authors upon written request.
Funding Sources
The CDD TB has been developed thanks to funding from the Bill and Melinda Gates Foundation (Grant#49852 “Collaborative drug discovery for TB through a novel database of SAR data optimized to promote data archiving and sharing”).
R.C.R. acknowledges the American Reinvestment and Recovery Act Grant 1RC1AI086677-01 that provided support for the presented study (National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID)) – “Targeting MDR-Tuberculosis.”
S.E. acknowledges that the earlier Bayesian models described and used herein were developed with support from Award Number R43 LM011152-01 “Biocomputation across distributed private datasets to enhance drug discovery” from the National Library of Medicine. TB Mobile was developed with funding from Award Number 2R42AI088893-02 “Identification of novel therapeutics for tuberculosis combining cheminformatics, diverse databases and logic based pathway analysis” from the National Institutes of Allergy and Infectious Diseases.
J.S.F. acknowledges funding from NIH/NIAID (2R42AI088893-02), Rutgers University–NJMS, and the Foundation of UMDNJ.
Footnotes
Supporting Information
Supplemental Tables 1 – 3
Supplemental Figures 1 – 5
This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of Interest
SE is a consultant for Collaborative Drug Discovery, Inc.
REFERENCES
- 1.Balganesh TS, Alzari PM, Cole ST. Rising standards for tuberculosis drug development. Trends Pharmacol Sci. 2008;29:576–581. doi: 10.1016/j.tips.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 3.Ballel L, Field RA, Duncan K, Young RJ. New small-molecule synthetic antimycobacterials. Antimicrob Agents Chemother. 2005;49:2153–2163. doi: 10.1128/AAC.49.6.2153-2163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd., Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469(7331):483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 6.Payne DA, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Disc. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 7.Wei JR, Krishnamoorthy V, Murphy K, Kim JH, Schnappinger D, Alber T, Sassetti CM, Rhee KY, Rubin EJ. Depletion of antibiotic targets has widely varying effects on growth. Proc Natl Acad Sci U S A. 2011;108(10):4176–4181. doi: 10.1073/pnas.1018301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maddry JA, Ananthan S, Goldman RC, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Reynolds RC, Secrist JA, 3rd., Sosa MI, White EL, Zhang W. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinb) 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ananthan S, Faaleolea ER, Goldman RC, Hobrath JV, Kwong CD, Laughon BE, Maddry JA, Mehta A, Rasmussen L, Reynolds RC, Secrist JA, 3rd., Shindo N, Showe DN, Sosa MI, Suling WJ, White EL. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds RC, Ananthan S, Faaleolea E, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Sosa MI, Thammasuvimol E, White EL, Zhang W, Secrist JA., 3rd. High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2012;92:72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RE, Protopopova M, Crooks E, Slayden RA, Terrot M, Barry CE., 3rd. Combinatorial lead optimization of [1,2]-diamines based on ethambutol as potential antituberculosis preclinical candidates. J Comb Chem. 2003;5(2):172–187. doi: 10.1021/cc020071p. [DOI] [PubMed] [Google Scholar]
- 12.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307(5707):223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 13.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DV, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011;10(3):188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 14.Prakash O, Ghosh I. Developing an antituberculosis compounds database and data mining in the search of a motif responsible for the activity of a diverse class of antituberculosis agents. J Chem Inf Model. 2006;46(1):17–23. doi: 10.1021/ci050115s. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Garcia A, Galvez J, de Julian-Ortiz JV, Garcia-Domenech R, Munoz C, Guna R, Borras R. Search of chemical scaffolds for novel antituberculosis agents. J Biomol Screen. 2005;10(3):206–214. doi: 10.1177/1087057104273486. [DOI] [PubMed] [Google Scholar]
- 16.Planche AS, Scotti MT, Lopez AG, de Paulo Emerenciano V, Perez EM, Uriarte E. Design of novel antituberculosis compounds using graph-theoretical and substructural approaches. Mol Divers. 2009;13(4):445–458. doi: 10.1007/s11030-009-9129-9. [DOI] [PubMed] [Google Scholar]
- 17.Prathipati P, Ma NL, Keller TH. Global Bayesian models for the prioritization of antitubercular agents. J Chem Inf Model. 2008;48(12):2362–2370. doi: 10.1021/ci800143n. [DOI] [PubMed] [Google Scholar]
- 18.Jones DR, Ekins S, Li L, Hall SD. Computational approaches that predict metabolic intermediate complex formation with CYP3A4 (+b5) Drug Metab Dispos. 2007;35(9):1466–1475. doi: 10.1124/dmd.106.014613. [DOI] [PubMed] [Google Scholar]
- 19.Ekins S, Freundlich JS. Validating new tuberculosis computational models with public whole cell screening aerobic activity datasets. Pharm Res. 2011;28:1859–1869. doi: 10.1007/s11095-011-0413-x. [DOI] [PubMed] [Google Scholar]
- 20.Ekins S, Kaneko T, Lipinksi CA, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Ernst S, Yang J, Goncharoff N, Hohman M, Bunin B. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis. Mol BioSyst. 2010;6:2316–2324. doi: 10.1039/c0mb00104j. [DOI] [PubMed] [Google Scholar]
- 21.Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin B. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- 22.Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models. PLOSONE. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekins S, Reynolds R, Kim H, Koo M-S, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian Models Leveraging Bioactivity and Cytotoxicity Information for Drug Discovery. Chem Biol. 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekins S, Freundlich JS, Hobrath JV, White EL, Reynolds RC. Combining Computational Methods for Hit to Lead Optimization in Mycobacterium tuberculosis Drug Discovery. Pharm Res. 2013 doi: 10.1007/s11095-013-1172-7. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzblau SG, DeGroote MA, Cho SH, Andries K, Nuermberger E, Orme IM, Mdluli K, Angulo-Barturen I, Dick T, Dartois V, Lenaerts AJ. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis (Edinb) 2012;92(6):453–488. doi: 10.1016/j.tube.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Anon Collaborative Drug Discovery, Inc. http://www.collaborativedrug.com/register.
- 27.Ekins S, Gupta RR, Gifford E, Bunin BA, Waller CL. Chemical space: missing pieces in cheminformatics. Pharm Res. 2010;27(10):2035–2039. doi: 10.1007/s11095-010-0229-0. [DOI] [PubMed] [Google Scholar]
- 28.Hohman M, Gregory K, Chibale K, Smith PJ, Ekins S, Bunin B. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Disc Today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Anon The PubChem Database. http://pubchem.ncbi.nlm.nih.gov/
- 30.Bender A, Scheiber J, Glick M, Davies JW, Azzaoui K, Hamon J, Urban L, Whitebread S, Jenkins JL. Analysis of Pharmacology Data and the Prediction of Adverse Drug Reactions and Off-Target Effects from Chemical Structure. ChemMedChem. 2007;2(6):861–873. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- 31.Klon AE, Lowrie JF, Diller DJ. Improved naive Bayesian modeling of numerical data for absorption, distribution, metabolism and excretion (ADME) property prediction. J Chem Inf Model. 2006;46(5):1945–1956. doi: 10.1021/ci0601315. [DOI] [PubMed] [Google Scholar]
- 32.Hassan M, Brown RD, Varma-O'brien S, Rogers D. Cheminformatics analysis and learning in a data pipelining environment. Mol Divers. 2006;10(3):283–299. doi: 10.1007/s11030-006-9041-5. [DOI] [PubMed] [Google Scholar]
- 33.Rogers D, Brown RD, Hahn M. Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J Biomol Screen. 2005;10(7):682–686. doi: 10.1177/1087057105281365. [DOI] [PubMed] [Google Scholar]
- 34.Anon R. http://www.r-project.org/
- 35.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, Gonzalez R, Lozano S, Huss S, Santos-Villarejo A, Martin-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuinan-Blanco M, Lavandera JL, Perez-Herran E, Gamo-Benito FJ, Garcia-Bustos JF, Barros D, Castro JP, Cammack N. Fueling Open-Source Drug Discovery: 177 Small-Molecule Leads against Tuberculosis. ChemMedChem. 2013;8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarker M, Talcott C, Madrid P, Chopra S, Bunin BA, Lamichhane G, Freundlich JS, Ekins S. Combining cheminformatics methods and pathway analysis to identify molecules with whole-cell activity against Mycobacterium tuberculosis. Pharm Res. 2012;29:2115–2127. doi: 10.1007/s11095-012-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekins S, Clark AM, Sarker M. TB Mobile: A Mobile App for Anti-tuberculosis Molecules with Known Targets. J Cheminform. 2013;5:13. doi: 10.1186/1758-2946-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geppert H, Vogt M, Bajorath J. Current trends in ligand-based virtual screening: molecular representations, data mining methods, new application areas, and performance evaluation. J Chem Inf Model. 2010;50(2):205–216. doi: 10.1021/ci900419k. [DOI] [PubMed] [Google Scholar]
- 39.Heikamp K, Bajorath J. Comparison of confirmed inactive and randomly selected compounds as negative training examples in support vector machine-based virtual screening. J Chem Inf Model. 2013;53(7):1595–1601. doi: 10.1021/ci4002712. [DOI] [PubMed] [Google Scholar]
- 40.Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, Fitzgerald M, Dai H, Zhang L, Hung DT. Identification of Novel Inhibitors of M. tuberculosis Growth Using Whole Cell Based High-Throughput Screening. ACS Chem Biol. 2012;7:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mak PA, Rao SP, Ping Tan M, Lin X, Chyba J, Tay J, Ng SH, Tan BH, Cherian J, Duraiswamy J, Bifani P, Lim V, Lee BH, Ling Ma N, Beer D, Thayalan P, Kuhen K, Chatterjee A, Supek F, Glynne R, Zheng J, Boshoff HI, Barry CE, 3rd., Dick T, Pethe K, Camacho LR. A High-Throughput Screen To Identify Inhibitors of ATP Homeostasis in Non-replicating Mycobacterium tuberculosis. ACS Chem Biol. 2012;7(7):1190–1197. doi: 10.1021/cb2004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnet S, Hartkoorn RC, Szekely R, Pato J, Triccas JA, Schneider P, Szantai-Kis C, Orfi L, Chambon M, Banfi D, Bueno M, Turcatti G, Keri G, Cole ST. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis (Edinb) 2010;90(6):354–360. doi: 10.1016/j.tube.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Cortes C, Vapnik V. Support vector networks. Machine Learn. 1995;20:273–293. [Google Scholar]
- 44.Chang CC, Lin CJ. LIBSVM: A library for support vector machines. 2001 [Google Scholar]
- 45.Bennet KP, Campbell C. Support vector machines: Hype or hallelujah. SIGKDD Explorations. 2000;2:1–13. [Google Scholar]
- 46.Brown MPS, Grundy WN, Lin D, Christianini N, Sugnet CW, Furey TS, Ares M, Jr., Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burbidge R, Trotter M, Buxton B, Holden S. drug design by machine learning: support vector machines for pharmaceutical analysis. Computers and Chemistry. 2001;26:5–14. doi: 10.1016/s0097-8485(01)00094-8. [DOI] [PubMed] [Google Scholar]
- 48.Cai Y-D, Liu X-J, Xu X-B, Chou K-C. Support vector machines for the classification and prediction of β-turn types. J Peptide Science. 2002;8:297–301. doi: 10.1002/psc.401. [DOI] [PubMed] [Google Scholar]
- 49.Kriegl JM, Arnhold T, Beck B, Fox T. A support vector machine approach to classify human cytochrome P450 3A4 inhibitors. J Comput Aided Mol Des. 2005;19(3):189–201. doi: 10.1007/s10822-005-3785-3. [DOI] [PubMed] [Google Scholar]
- 50.Hammann F, Gutmann H, Baumann U, Helma C, Drewe J. Classification of cytochrome p(450) activities using machine learning methods. Mol Pharm. 2009;6(6):1920–1926. doi: 10.1021/mp900217x. [DOI] [PubMed] [Google Scholar]
- 51.Bikadi Z, Hazai I, Malik D, Jemnitz K, Veres Z, Hari P, Ni Z, Loo TW, Clarke DM, Hazai E, Mao Q. Predicting P-glycoprotein-mediated drug transport based on support vector machine and three-dimensional crystal structure of P-glycoprotein. PLoS One. 2011;6(10):e25815. doi: 10.1371/journal.pone.0025815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen K, Mika S, Schroeter T, Sutter A, ter Laak A, Steger-Hartmann T, Heinrich N, Muller KR. Benchmark data set for in silico prediction of Ames mutagenicity. J Chem Inf Model. 2009;49(9):2077–2081. doi: 10.1021/ci900161g. [DOI] [PubMed] [Google Scholar]
- 53.Lombardo F, Obach RS, Dicapua FM, Bakken GA, Lu J, Potter DM, Gao F, Miller MD, Zhang Y. A hybrid mixture discriminant analysis-random forest computational model for the prediction of volume of distribution of drugs in human. J Med Chem. 2006;49(7):2262–2267. doi: 10.1021/jm050200r. [DOI] [PubMed] [Google Scholar]
- 54.Liaw A, Wiener M. Classification and regression by random forest. R News. 2002;2/3:18–22. [Google Scholar]
- 55.Solimeo R, Zhang J, Kim M, Sedykh A, Zhu H. Predicting chemical ocular toxicity using a combinatorial QSAR approach. Chem Res Toxicol. 2012;25(12):2763–2769. doi: 10.1021/tx300393v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arimoto R, Prasad MA, Gifford EM. Development of CYP3A4 inhibition models: comparisons of machine-learning techniques and molecular descriptors. J Biomol Screen. 2005;10(3):197–205. doi: 10.1177/1087057104274091. [DOI] [PubMed] [Google Scholar]
- 57.Zientek M, Stoner C, Ayscue R, Klug-McLeod J, Jiang Y, West M, Collins C, Ekins S. Integrated in silico-in vitro strategy for addressing cytochrome P450 3A4 time-dependent inhibition. Chem Res Toxicol. 2010;23(3):664–676. doi: 10.1021/tx900417f. [DOI] [PubMed] [Google Scholar]
- 58.Ekins S, Williams AJ, Xu JJ. A Predictive Ligand-Based Bayesian Model for Human Drug Induced Liver Injury. Drug Metab Dispos. 2010;38:2302–2308. doi: 10.1124/dmd.110.035113. [DOI] [PubMed] [Google Scholar]
- 59.Astorga B, Ekins S, Morales M, Wright SH. Molecular Determinants of Ligand Selectivity for the Human Multidrug And Toxin Extrusion Proteins, MATE1 and MATE-2K. J Pharmacol Exp Ther. 2012;341(3):743–755. doi: 10.1124/jpet.112.191577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong Z, Ekins S, Polli JE. Structure-activity relationship for FDA approved drugs as inhibitors of the human sodium taurocholate cotransporting polypeptide (NTCP) Mol Pharm. 2013;10(3):1008–1019. doi: 10.1021/mp300453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan Y, Li L, Kim G, Ekins S, Wang H, Swaan PW. Identification and Validation of Novel hPXR Activators Amongst Prescribed Drugs via Ligand-Based Virtual Screening. Drug Metab Dispos. 2011;39:337–344. doi: 10.1124/dmd.110.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langdon SR, Mulgrew J, Paolini GV, van Hoorn WP. Predicting cytotoxicity from heterogeneous data sources with Bayesian learning. J Cheminform. 2010;2(1):11. doi: 10.1186/1758-2946-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broccatelli F, Mannhold R, Moriconi A, Giuli S, Carosati E. QSAR Modeling and Data Mining Link Torsades de Pointes Risk to the Interplay of Extent of Metabolism, Active Transport, and hERG Liability. Mol Pharm. 2013 doi: 10.1021/mp300156r. [DOI] [PubMed] [Google Scholar]
- 64.Xu C, Cheng F, Chen L, Du Z, Li W, Liu G, Lee PW, Tang Y. In silico prediction of chemical Ames mutagenicity. J Chem Inf Model. 2012;52(11):2840–2847. doi: 10.1021/ci300400a. [DOI] [PubMed] [Google Scholar]
- 65.Ekins S, Freundlich JS. Computational models for tuberculosis drug discovery. Methods Mol Biol. 2013;993:245–262. doi: 10.1007/978-1-62703-342-8_16. [DOI] [PubMed] [Google Scholar]
- 66.Ekins S, Freundlich JS, Choi I, Sarker M, Talcott C. Computational Databases, Pathway and Cheminformatics Tools for Tuberculosis Drug Discovery. Trends in Microbiology. 2011;19:65–74. doi: 10.1016/j.tim.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Periwal V, Kishtapuram S, Consortium OS, Scaria V. Computational models for in-vitro anti-tubercular activity of molecules based on high-throughput chemical biology screening datasets. BMC Pharmacol. 2012;12(1):1. doi: 10.1186/1471-2210-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Periwal V, Rajappan JK, Jaleel AU, Scaria V. Predictive models for anti-tubercular molecules using machine learning on high-throughput biological screening datasets. BMC Res Notes. 2011;4:504. doi: 10.1186/1756-0500-4-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gamo F-J, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera J-L, Vanderwall DE, Green DVS, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 70.Ekins S, Williams AJ. When Pharmaceutical Companies Publish Large Datasets: An Abundance Of Riches Or Fool’s Gold. Drug Disc Today. 2010;15:812–815. doi: 10.1016/j.drudis.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 71.Seal A, Yogeeswari P, Sriram D, Consortium O, Wild DJ. Enhanced ranking of PknB Inhibitors using data fusion methods. J Cheminform. 2013;5(1):2. doi: 10.1186/1758-2946-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swamidass SJ, Azencott CA, Daily K, Baldi P. A CROC stronger than ROC: measuring, visualizing and optimizing early retrieval. Bioinformatics. 2010;26(10):1348–1356. doi: 10.1093/bioinformatics/btq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. Rapid Identification of P-glycoprotein Substrates and Inhibitors. Drug Metab Dispos. 2006;34:1976–1984. doi: 10.1124/dmd.106.012351. [DOI] [PubMed] [Google Scholar]
- 74.Guner OF, Henry DR. Metric for analyzing hit lists and pharmacophores. In: Guner OF, editor. Pharmacophore perception, development, and use in drug design. La Jolla, CA: International University Line; 2000. pp. 191–211. [Google Scholar]
- 75.Liew CY, Lim YC, Yap CW. Mixed learning algorithms and features ensemble in hepatotoxicity prediction. J Comput Aided Mol Des. 2011;25(9):855–871. doi: 10.1007/s10822-011-9468-3. [DOI] [PubMed] [Google Scholar]
- 76.Willett P. Combination of similarity rankings using data fusion. J Chem Inf Model. 2013;53(1):1–10. doi: 10.1021/ci300547g. [DOI] [PubMed] [Google Scholar]
- 77.Rodgers AD, Zhu H, Fourches D, Rusyn I, Tropsha A. Modeling liver-related adverse effects of drugs using knearest neighbor quantitative structure-activity relationship method. Chem Res Toxicol. 2010;23(4):724–732. doi: 10.1021/tx900451r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Embrechts MJ, Ekins S. Classification of Metabolites with Kernel-Partial Least Squares (K-PLS) Drug Metab Dispos. 2007;35(3):325–327. doi: 10.1124/dmd.106.013185. [DOI] [PubMed] [Google Scholar]
- 79.Ivanenkov YA, Savchuk NP, Ekins S, Balakin KV. Computational mapping tools for drug discovery. Drug Discov Today. 2009 doi: 10.1016/j.drudis.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 80.Spjuth O, Carlsson L, Alvarsson J, Georgiev V, Willighagen E, Eklund M. Open source drug discovery with bioclipse. Curr Top Med Chem. 2012;12(18):1980–1986. doi: 10.2174/156802612804910287. [DOI] [PubMed] [Google Scholar]
- 81.Spjuth O, Willighagen EL, Guha R, Eklund M, Wikberg JE. Towards interoperable and reproducible QSAR analyses: Exchange of datasets. J Cheminform. 2010;2(1):5. doi: 10.1186/1758-2946-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spjuth O, Helmus T, Willighagen EL, Kuhn S, Eklund M, Wagener J, Murray-Rust P, Steinbeck C, Wikberg JE. Bioclipse: an open source workbench for chemo- and bioinformatics. BMC Bioinformatics. 2007;8:59. doi: 10.1186/1471-2105-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walker T, Grulke CM, Pozefsky D, Tropsha A. Chembench: a cheminformatics workbench. Bioinformatics. 2010;26(23):3000–3001. doi: 10.1093/bioinformatics/btq556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ekins S, Bunin BA. The Collaborative Drug Discovery (CDD) database. Methods Mol Biol. 2013;993:139–154. doi: 10.1007/978-1-62703-342-8_10. [DOI] [PubMed] [Google Scholar]
- 85.Gupta RR, Gifford EM, Liston T, Waller CL, Bunin B, Ekins S. Using open source computational tools for predicting human metabolic stability and additional ADME/TOX properties. Drug Metab Dispos. 2010;38:2083–2090. doi: 10.1124/dmd.110.034918. [DOI] [PubMed] [Google Scholar]
- 86.Ponder EL, Freundlich JS, Sarker M, Ekins S. Computational Models For Neglected Diseases: Gaps and Opportunities. Pharm Res. 2013 doi: 10.1007/s11095-013-1170-9. In Press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.