Figure 1.
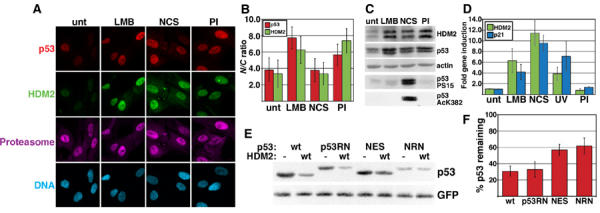
p53 can be degraded in the nucleus. (A) p53, HDM2, and proteasome subcellular localization in normal human fibroblasts (WS1). Cells were left untreated (unt), or treated with 20 nM LMB for 6 h, 640 ng/ml NCS for 4 h, or PIs for 4 h, and stained with antibodies to p53, HDM2, and the proteasome. (B) p53 and HDM2 nuclear:cytoplasmic (N/C) ratios for immunofluorescently stained WS1. See Materials and methods for determination of p53 and HDM2 N/C ratios. p53 ratios are depicted by gray bars and HDM2 ratios by white bars. Values represent the mean (±s.d.) of >30 cells. (C) Immunoblots of p53, HDM2, p53 phosphorylated S15 (PS15), and p53 acetylated lysine 382 (AcK382). WS1 cells were treated as in (A), then immunoblotted with antibodies to p53 (DO-1), HDM2 (IF2), and actin. The membrane was stripped and re-probed with an antibody to p53 PS15. Separate samples were lysed in acetylation lysis buffer, then stained with an antibody to AcK382. (D) p53 target gene induction. cDNA was harvested from WS1 that were untreated; LMB, NCS, or PI-treated (see (A)); or UV irradiated. HDM2 (white bars) or p21 (gray bars) gene induction was measured by real-time quantitative PCR. Induction is expressed as fold induction relative to untreated. Error bars indicate the 95% CI of ≥3 experiments. (E) Degradation of cytoplasmic p53 by HDM2. 2KO or SaOS-2 cells (data not shown) were transfected with plasmids containing luciferase, GFP, 150 ng p53 (wt, p53RN, NES, or NRN), and 1.2 μg HDM2 (wt) or empty vector (pcDNA3, (−)). Equal luciferase units of transfected cell lysates were immunoblotted with antibodies to p53 or GFP. (F) Densitometric analysis of p53 Western blots. Percent p53 remaining was calculated as the ratio of p53 co-transfected with wt HDM2 to p53, with empty vector normalized to GFP. Bars represent the mean band intensity±1 s.d. for >3 experiments.
