Figure 1.
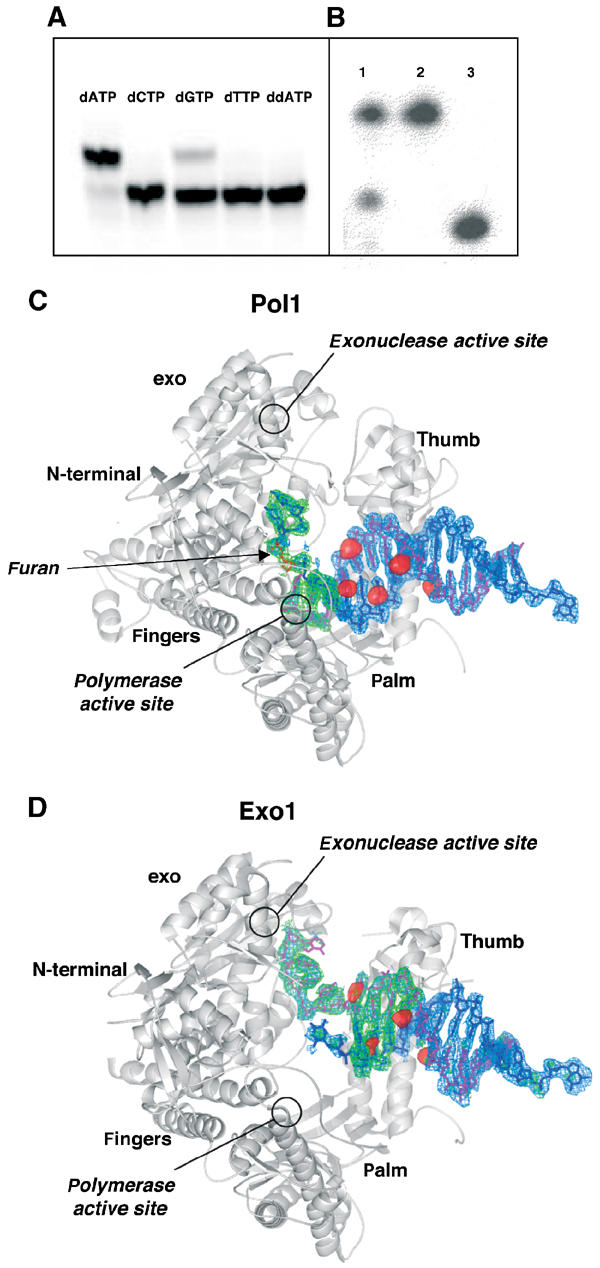
Crystallization of RB69 gp43 exo− with furan-containing DNA. (A) RB69 gp43 exo− is able to extend a primer by incorporating dAMP, and to a lesser extent dGMP, opposite an abasic site. No measurable incorporation of the pyrimidines was detected nor was incorporation of ddATP. (B) dATP is incorporated by RB69 gp43 exo− under crystallization conditions. The primer has been extended by one base and there is no evidence of residual exonuclease activity (lane 1: dissolved crystal; lane 2: template; lane 3: primer). (C) Molecule with DNA in the polymerase active site (Pol1). An Fo–Fc map (blue) calculated with DNA omitted and contoured to 3σ and an Fo–Fc simulated annealing omit map (green) with a 10 Å sphere omitted around the abasic site and contoured to 3.5σ are superimposed on the protein model. The red surfaces represent peaks in the anomalous difference Fourier map calculated from the brominated DNA data contoured at 5σ. The primer strand is shown in magenta and the abasic site in red. (D) Molecule with DNA in the exonuclease active site (Exo1). All maps are as those in (C) with the exception that the simulated annealing omit maps were calculated with a 10 Å sphere omitted around both the 3′ end of the primer and the 5′ end of the template. Note that the 5′ end of the template becomes disordered once it disassociates from the primer, and thus the furan is not visible in this structure nor is the 5′ single-stranded template. The DNA sequence used in co-crystallization is described in Materials and methods.
