Figure 3.
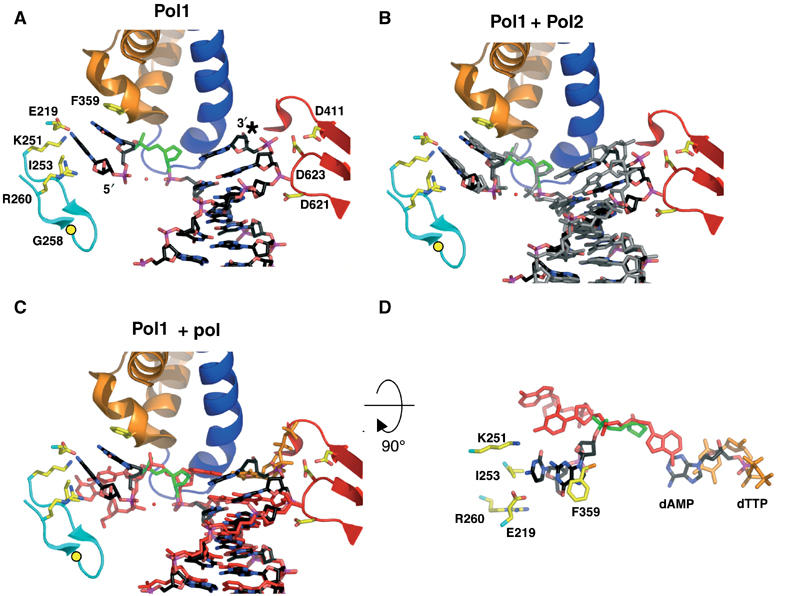
Furan-containing DNA in the polymerase active site is distorted and differs significantly from normal DNA. (A) The polymerase active site of Pol1. The protein backbone is colored according to the different protein domains: palm (red), fingers (blue), exonuclease (cyan) and N-terminal (orange). The furan moiety is in green and the dAMP incorporated opposite the furan is marked with an asterisk. Gly258 is shown as a yellow circle and the red sphere is a modeled water molecule. (B) DNA from Pol2 (gray) is superimposed on the Pol1 model after alignment via their palm domains (residues 383–468 and 573–729). (C) Superposition of DNA from the ternary complex (red; Franklin et al, 2001) with Pol1 (black) shows marked differences in the DNA conformation. Note that the last base pair in the Pol1 binary complex superimposes with the ultimate base pair in the ternary complex, composed of the incoming dTTP (orange) and its template base, which implies that the DNA has not translocated in the Pol1 binary complex. (D) The DNA from (C) is rotated 90° toward the reader and with most protein residues removed from view for clarity. The trajectory of the 5′ template DNA from Pol1 (black) differs from that seen in the ternary complex (red). As in (C), the furan moiety from Pol1 is shown in green and the incoming dTTP from the ternary complex model in orange.
