Figure 5.
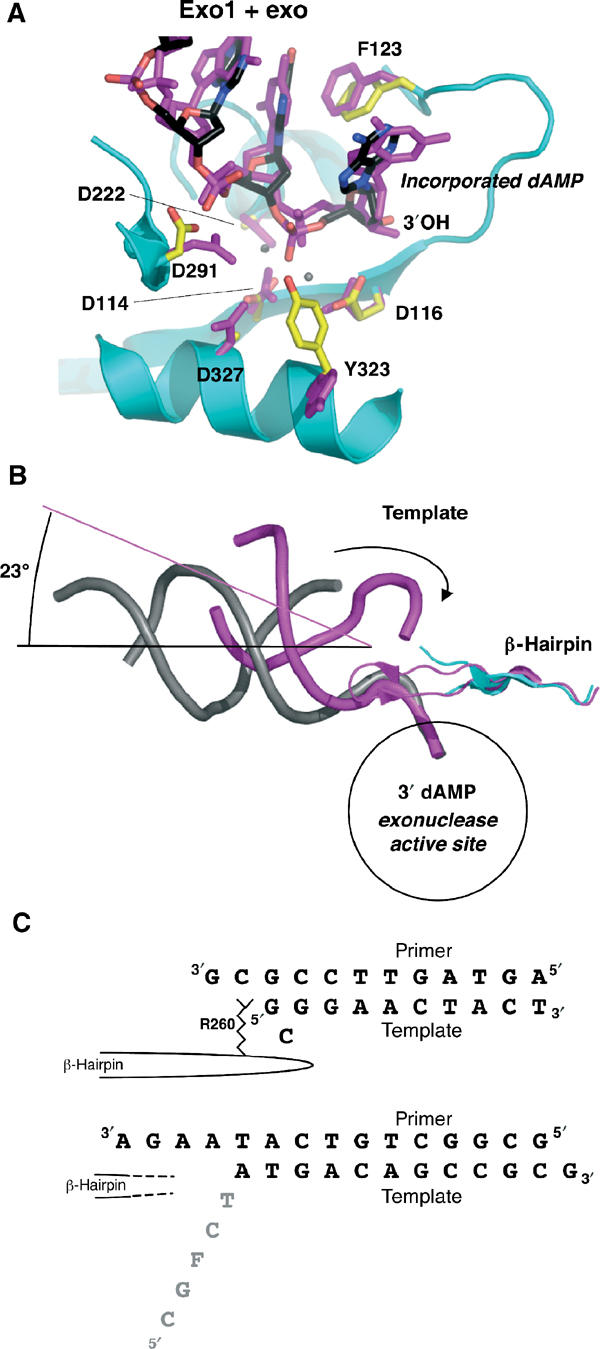
DNA in the exonuclease active site. (A) Exo1 model (protein: cyan; DNA: black) aligned with exo complex (magenta, with calcium ions shown as gray spheres; Shamoo and Steitz, 1999). The incorporated dAMP opposite the furan at the primer terminus is flipped into the exonuclease domain. The position of the single-stranded DNA and its contacts with the protein are similar between the model with undamaged DNA and that with furan-containing DNA. (B) Same alignment as in (A), showing the entire length of the DNA backbone (Exo1 in gray, exo model in magenta). The template (indicated by the arrow) in the exo model is in contact with the β-hairpin loop. In Exo1, the unpaired portion of the template is disordered. The β-hairpin loop is in a similar position in both models, but the tip of the loop is disordered in Exo1. The helical axes are shown to indicate the angular difference between the two DNA molecules. (C) Comparison of the DNA sequences in the exo model (Shamoo and Steitz, 1999) top sequence, and Exo1, bottom sequence. In the exo model, the Arg260 in the β-hairpin loop stacks against the template strand, which forms a G–G base pair and a bulged, unpaired C. In contrast, the tip of the β-hairpin loop is disordered in Exo1, and so is the single-stranded template (gray).
