Figure 6.
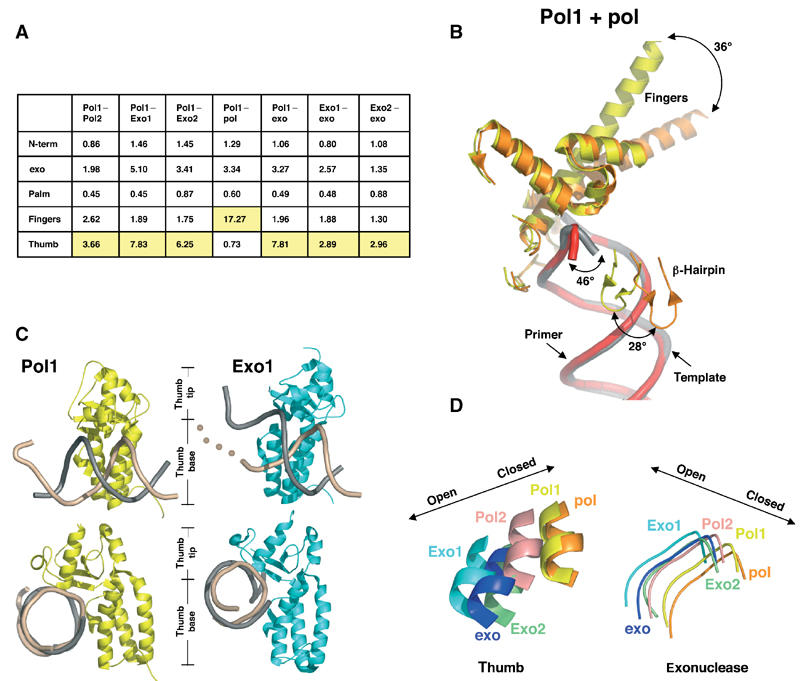
Protein domain movements. (A) Table of root mean square (r.m.s.) deviations (Å). The largest deviations between pairs of conformations are highlighted in yellow. Molecules were aligned using the palm residues (383–468 and 573–729). (B) Protein movements between Pol1 (yellow, with DNA in gray) and ternary complex (orange, with DNA in red). The largest protein movement (36°) is seen in the fingers domain. (C) Thumb position in Pol1 and Exo1. The thumb domains from Pol1 (yellow) and Exo1 (cyan) contact template (beige) and primer (gray) DNA. The beige dots illustrate that the template DNA in Exo1 has no visible density once it unwinds from the primer. Most of the contacts between the thumb and the DNA are maintained between the polymerizing and editing modes, suggesting that the DNA has not translocated during the switch from the polymerase to exonuclease active sites. The bottom set of images have been oriented to look down the DNA helical axis to show the increased opening of the thumb tip in the Exo1 model. (D) Coordinated motion of the thumb and exonuclease domains. All molecules have been aligned via the palm domain (residues 383–468 and 573–729). A loop from the exonuclease domain (residues 120–129) and a portion of helix W from the thumb domain (residues 809–815) are shown. Movement of these domains to the left represents a more open conformation as described for the apo and exo models, and movement to the right represents a closed conformation as described for the ternary complex. A coordination between exonuclease and thumb movements exists, such that as the thumb relaxes its grip the exonuclease domain shifts closer to the thumb.
