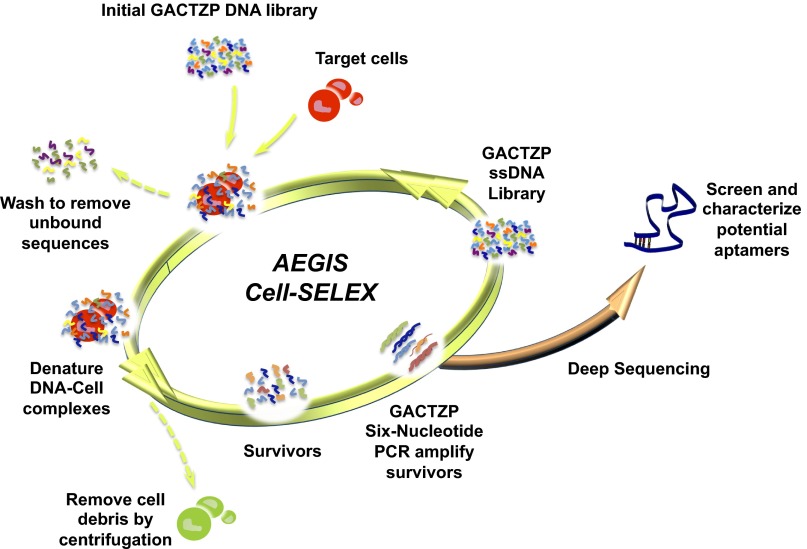
Fig. 2.
Schematic of the AEGIS cell–SELEX used here. A GACTZP DNA library, consisting of randomized sequences and primer sites, was first incubated with the target cells. Unbound sequences were then removed by washing. Bound sequences having affinity to the target cells were collected. “Survivors” in the enriched collection were amplified using a FITC-labeled 5′-primer and a biotinylated 3′-primer by GACTZP six-nucleotide PCR (22). Enriched FITC-conjugated single-stranded DNA obtained from PCR products was used for the next round of selection. The binding affinity of survivors from 9th round up to 12th round of selection was monitored by flow cytometer. The survivors from 12th round selection were subjected to deep sequencing.