Figure 2.
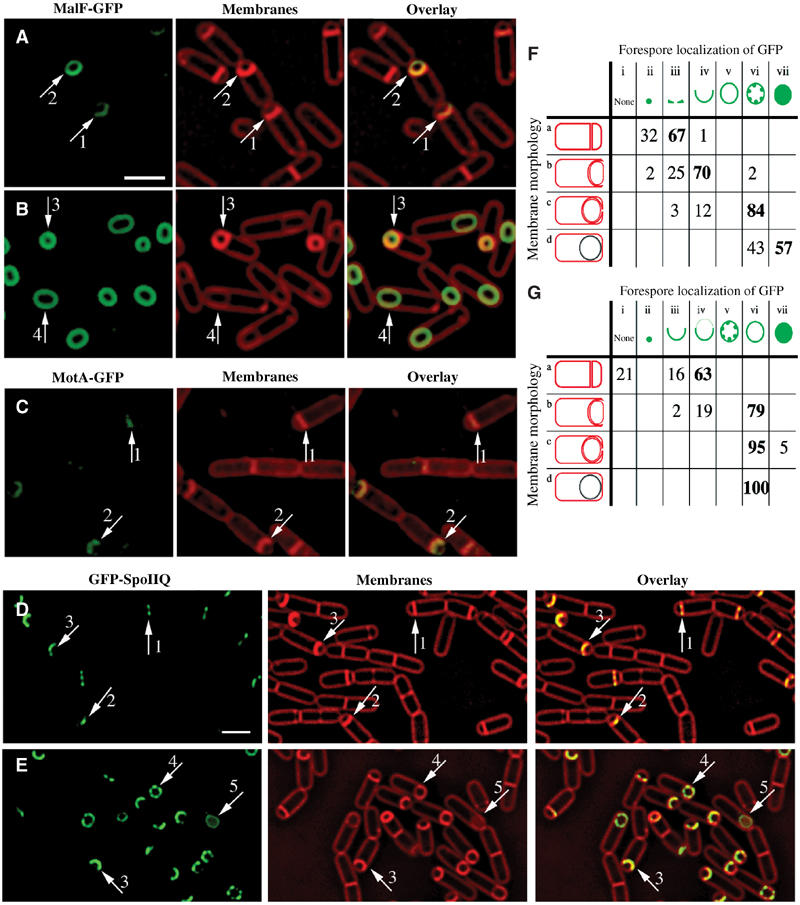
Localization patterns of forespore-expressed heterologous membrane proteins and the native forespore protein SpoIIQ. (A, B) MalF-GFP (green) 2 h (A, hereafter t2) and 3 h (B, hereafter t3) after the onset of sporulation stained with FM 4-64 (red). In most sporangia, engulfment is underway at t2 and complete by t3. (C) MotA-GFP (green) at t2 stained with Mitotracker Red (red). In panels A and C, arrows 1 and 2 represent sporangia with flat or slightly curving polar septa. In panel B, arrows 3 and 4 represent sporangia before and after membrane fusion, respectively. (D) GFP-SpoIIQ (green) at t2 and (E) t3 stained with FM 4-64 (red). In panels D and E, arrows 1, 2 and 3 represent early sporangia and arrows 4 and 5 represent sporangia late in engulfment. The scale bar in (A, D) is 2 μm. (F, G) Quantitation of GFP localization at various stages of engulfment (a–d). Numbers refer to the percentage of sporangia in each engulfment class with the indicated localization pattern; those in bold represent the predominant class. (F) GFP-SpoIIQ (AR126) localization. Seven subclasses of GFP localization were scored: (i) none; (ii) medial focus; (iii) ring; (iv) tracking mother cell engulfing membrane; (v) freely diffusible; (vi) punctate; (vii) soluble. A total of 504 sporangia were scored from t2 and t3. (G) MalF-GFP (AR4) localization. Seven subclasses for GFP localization were scored: (i) none; (ii) focus formation; (iii) tracking mother cell engulfing membrane; (iv) enriched at septum but ahead of engulfing membranes; (v) punctate; (vi) freely diffusible; (vii) soluble. A total of 296 sporangia were scored from t2 and t3.
