Abstract
The emergence of carbapenemase-producing bacteria poses a new challenge in the management of antibiotic therapies for patients. This report describes a new method using ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) for rapid detection of carbapenemase activity in enterobacteria, Pseudomonas aeruginosa, and Acinetobacter baumannii. In a panel of 78 isolates, including 41 carbapenemase-producing strains, the ULPC-MS/MS assay showed 100% agreement with molecular characterization, whereas six carbapenemase-producing isolates were not detected by the modified Hodge test.
TEXT
The emergence and dissemination of resistance to carbapenem in Gram-negative pathogens such as Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa represent a challenge for optimizing antibiotic therapies and preventing outbreaks, especially regarding carbapenemase-producing bacteria, by comparison to other resistance mechanisms. Using the Ambler classification of beta-lactamases, the principal carbapenemase enzymes detected in infected patients worldwide belong to class B (metallo-beta-lactamases, including IMP, VIM, and NDM), to class A (beta-lactamases, including Klebsiella pneumoniae carbapenemase [KPC]), and to class D (oxacillinases, including OXA-48 and OXA-23). Moreover, clinical infections due to these carbapenemase-producing strains, which are also resistant to most beta-lactam antibiotics, are associated with higher morbidity and mortality (1). Bacterial resistance to carbapenem is usually screened using susceptibility and phenotypic assays such as the modified Hodge test (MHT) (2). However, these tests have shown poor sensitivity and specificity in detecting carbapenemase-producing isolates, especially in A. baumannii (3). While PCR-based detection of the carbapenemase gene is considered the gold standard method, in view of the high genetic diversity of genes coding for carbapenemase, numerous PCR assays targeting the most prevalent enzymes must be developed in this approach (2, 4). Recently, mass spectrometry (MS) technologies, such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS), have been shown to be capable of characterizing carbapenemase-producing bacteria (5, 6, 7, 8).
This study reports on a rapid UPLC-MS/MS assay to detect carbapenemase-producing Enterobacteriaceae, P. aeruginosa, and A. baumannii isolates based on the ability of bacterial isolates to hydrolyze ertapenem and meropenem in solution.
A total of 78 bacterial strains (27 reference strains and 51 strains isolated from clinical samples) were included in the study: 41 carbapenemase-producing strains (including 23 enterobacterial strains, 3 P. aeruginosa strains, and 15 A. baumannii strains) and 37 strains susceptible to carbapenems or harboring decreased susceptibility to carbapenems due to non-carbapenemase-based mechanisms. Escherichia coli ATCC 25922 and Citrobacter freundii NDM-1-positive strains were used as the negative and positive controls, respectively. All isolates were tested using imipenem, meropenem, and ertapenem MICs (Etest; bioMérieux, Marcy l'Etoile, France) and by the MHT following the CLSI recommendations (9). The molecular mechanism involved in carbapenem resistance was determined by PCR assays targeting blaNDM-1, blaVIM, blaIMP, blaKPC, blaOXA-48, blaOXA40, and blaOXA23 genes as previously described (10, 11, 12, 13). All carbapenemase-producing clinical strains were sent to the French Reference Centre on Emerging Antibiotic Resistance (Paris, France) for molecular characterization and clone determination. In addition, each strain was also subjected to the UPLC-MS/MS assay, which measures carbapenem hydrolysis in a solution spiked with the test bacterial strain. Briefly, the strains were cultured overnight on Mueller-Hinton agar (bioMérieux, La Balme Les Grottes, France) and the bacteria were dissolved in normal saline solution and adjusted to McFarland standard 4. A 1-ml volume of this suspension was incubated with 5 mg/ml of ertapenem in a first tube (TRC Products, Molsheim, France) and with 5 mg/ml of meropenem in a second tube (TRC Products) for 3 or 4 h at 36°C under conditions of smooth agitation. The suspensions were then centrifuged at 12,000 × g for 5 min, and 300 μl of the supernatant was mixed with 700 μl of methanol containing an internal standard (meropenem d6; TRC Products). The suspensions were centrifuged again at 12,000 × g for 5 min, and 200 μl of the supernatant was mixed with 800 μl of water. The concentrations of ertapenem and meropenem were measured using a UPLC-MS/MS system comprising an Acquity UPLC apparatus (Waters, Milford, MA) coupled to an Acquity tandem quadrupole detector (TQD) (Waters). Briefly, the chromatography step was performed using a BEH C18 column (Waters) eluted with mobile phases A (0.1% formic acid in acetonitrile) and B (0.1% formic acid in methanol), with an initial composition of 5% solution B. The volume of injection was 5 μl. The flow rate was 1 ml/min, and a 5% to 95% gradient was monitored for 1.5 min. Under these conditions, ertapenem and meropenem were eluted after 0.97 min and 0.48 min, respectively. The detection step by tandem mass spectrometry was done using an electrospray ionization interface in positive multiple-reaction-monitoring mode. Optimized multiple-reaction-monitoring settings include cone voltage (24 V for ertapenem and 18 V for meropenem) and collision energy (26 eV for ertapenem and 16 eV for meropenem). The transitions of [M-H]+ precursor ion products were a 476.0 to 114.0 m/z ratio for ertapenem, 383.9 to 254.1 m/z for meropenem, and 390.1 to 147.1 m/z for the internal standard (meropenem D6) (Fig. 1). Quantification of ertapenem and meropenem was done using the MassLynx software package (v4.0; Waters). The calibration curve for the matrix was linear over a concentration range of 0.5 to 10 mg/liter with a correlation coefficient value greater than 0.9. The rate of hydrolysis of ertapenem and meropenem was calculated by dividing the carbapenem concentration after the incubation step by the preincubation carbapenem concentration. Preliminary kinetic studies of ertapenem and meropenem degradation were performed for 5 carbapenem-resistant strains. Although hydrolysis of carbapenem in solution occurs within 3 h for NDM-1- and KPC-producing enterobacteria, the incubation time had to be increased to 4 h in order to detect the carbapenemase activity of one CHDL-producing strain of A. baumannii, one VIM-producing strain of P. aeruginosa, and one OXA-48-producing strain of K. pneumoniae (data not shown).
FIG 1.
UPLC-MS/MS chromatograms for ertapenem and meropenem (standard solution) for strains E. coli ATCC 25922 (left panels) and E. coli OXA-48 producer (right panels).
The results of the UPLC-MS/MS assays for the 78 isolates showed 100% agreement with molecular characterization (Table 1) and were consistent with the results published by Carvalhaes et al. using 73 strains, including SPM-1-producing P. aeruginosa and CHDL-producing A. baumannii (8). For the 37 non-carbapenemase-producing strains, the mean hydrolysis rates were 2.3% (range, 0% to 17%) and 1.9% (range, 0% to 10.4%) for meropenem and ertapenem, respectively; by comparison, for the 41 carbapenemase-producing strains, the mean hydrolysis rates were 84.6% (range, 38.3% to 100%) and 88.0% (range, 45.5% to 100%), respectively (P < 0.0001, Mann-Whitney test) (Fig. 2). The rate of ertapenem or meropenem hydrolysis was greater than 30% for all carbapenemase-producing strains. The natural oxacillinase OXA-51 of A. baumannii was not detected by the UPLC-MS/MS, whereas overproduction of this enzyme leading to decreased carbapenem efficacy was unambiguously detected (Table 1). In contrast, none of non-carbapenemase-producing isolates were able to hydrolyze ertapenem or meropenem at a rate of greater than 20%, even in multidrug-resistant (MDR) strains with overproduction of cephalosporinases or extended-spectrum β-lactamases. Accordingly, strains yielding carbapenem hydrolysis rates greater than 20% are tested using molecular assays in our routine practice. The UPLC-MS/MS method was able to detect all carbapenemase-producing strains (sensitivity, 100%; 95% confidence interval [CI], 91.4% to 100%), whereas only 32 of 41 (sensitivity, 78.1%; 95% CI, 62.4% to 89.4%) carbapenemase-producing strains were detected by MHT (P < 0.0046, chi-square test), a finding consistent with previous studies reporting both false-positive and false-negative results for MHT (3, 14). In addition, the correlation between the MIC and the hydrolysis rate was statistically significant, with Pearson coefficient values of 0.57 (95% CI, 0.39 to 0.70; P < 0.0001) and 0.52 (95% CI, 0.34 to 0.67; P < 0.0001) for ertapenem and meropenem, respectively. This phenotypic approach could be used in predicting the efficacy of carbapenems when combined with other active antibiotics as suggested by Tumbarello et al. (15). This correlation should be studied more closely in further studies both by increasing the amount of carbapenem in the UPLC MS/MS assay and by determining the carbapenem MICs in a larger range.
TABLE 1.
Rate of hydrolysis of ertapenem and meropenem for 78 strains of Gram-negative bacilli by MIC of carbapenem, results of modified Hodge test, and the molecular mechanism of carbapenemase-producing isolates
| Enzyme | Species (no. of strains) | No. of isolates with positive modified Hodge test result | MIC (range) (mg/liter) |
UPLC hydrolysis rate [% (range)] |
|||
|---|---|---|---|---|---|---|---|
| Imipenem | Ertapenem | Meropenem | Ertapenem | Meropenem | |||
| Carbapenemase-producing strains (n = 41) | |||||||
| KPC | Klebsiella pneumoniae (2) | 2 | 1 to >32 | 4 to >32 | 2 to >32 | 100 | 100 |
| IMP | Escherichia coli (1) | 1 | 1 | 0.5 | 0.5 | 100 | 100 |
| Pseudomonas aeruginosa (1) | 1 | >32 | >32 | >32 | 100 | 100 | |
| VIM | Pseudomonas aeruginosa (2) | 0 | >32 | >32 | >32 | 44 to 100 | 98 to 100 |
| Klebsiella pneumoniae (1) | 1 | 8 | 3 | 1 | 25 | 51 | |
| NDM-1 | Citrobacter freundii (1) | 1 | 8 | >32 | 12 | 100 | 100 |
| Acinetobacter baumannii (1) | 0 | >32 | >32 | >32 | 63 | 78 | |
| Klebsiella pneumoniae (2) | 1 | 16 to >32 | >32 | 6 to >32 | 97 to 100 | 100 | |
| Escherichia coli (1) | 0 | 1.5 | 1.5 | 0 | 89 | 91 | |
| OXA-48 | Klebsiella pneumoniae (6) | 6 | 0.75 to >32 | 1 to >32 | 0.75 to 16 | 98 to 100 | 99 to 100 |
| Escherichia coli (5) | 5 | 1.5 to >32 | 1.5 to >32 | 0.75 to >32 | 99 to 100 | 99 to 100 | |
| Citrobacter freundii (2) | 2 | 1 to 3 | 1.5 to 4 | 0.38 to 2 | 96 to 100 | 100 | |
| Serratia marcescens (2) | 2 | 1.5 to 16 | 1.5 to 16 | 0.75 to 8 | 100 | 100 | |
| OXA-23 | Acinetobacter baumannii (10) | 8 | 24 to >32 | 32 to >32 | 32 to >32 | 38 to >100 | 46 to >100 |
| OXA-40 | Acinetobacter baumannii (2) | 1 | >32 | >32 | >32 | 69 to 93 | 70 to 93 |
| OXA-51a | Acinetobacter baumannii (1) | 1 | >32 | >32 | >32 | 69 | 69 |
| OXA-58 | Acinetobacter baumannii (1) | 0 | 2 | 12 | 1.5 | 93 | 92 |
| MDRb non-carbapenemase-producing strains (n = 22) | Citrobacter freundii (1) | 0 | 0.125 | 0.008 | 0.032 | 0 | 0 |
| Enterobacter cloacae (4) | 0 | 0.38 to 32 | 0.19 to >32 | 0.19 to 12 | 0 to 11 | 0 to 8 | |
| Escherichia coli (11) | 0 | 0.125 to 1 | 0.008 to 16 | 0.012 to 4 | 0 to 15 | 0 to 4 | |
| Klebsiella pneumoniae (5) | 0 | 0.125 to 0.5 | 0.094 to 3 | 0.016 to 0.38 | 0 to 17 | 0 to 4 | |
| Klebsiella oxytoca (1) | 0 | 0.125 | 0.016 | 0.023 | 0 | 0 | |
| Susceptible strains (n = 15) | Escherichia coli (5) | 0 | 0.016 to 0.25 | 0.008 to 0.032 | 0.008 to 0.19 | 0 to 2 | 0 to 9 |
| Citrobacter freundii (2) | 0 | 0.38 | 0.016 to 0.19 | 0.032 to 0.064 | 0 | 0 to 6 | |
| Klebsiella pneumoniae (2) | 0 | 0.125 to 0.19 | 0.008 to 0.016 | 0.032 | 0 | 2 to 10 | |
| Klebsiella oxytoca (1) | 0 | 0.19 | 0.008 | 0.032 | 0 | 0 | |
| Pseudomonas aeruginosa (2) | 0 | 0.15 to 2 | 0.23 to 12 | 0.047 to 0.5 | 0 to 1 | 0 to 8 | |
| E. cloacae (2) | 0 | 0.19 to 0.25 | 0.032 to 0.5 | 0.023 to 0.38 | 0 to 5 | 0 to 5 | |
| Acinetobacter baumannii (1)c | 0 | 0.38 | 3 | 1.5 | 0 | 0 | |
Acinetobacter baumannii carrying overproduced OXA-51.
MDR, multidrug resistant.
Acinetobacter baumannii ATCC 19606 carrying the natural oxacillinase OXA-51.
FIG 2.
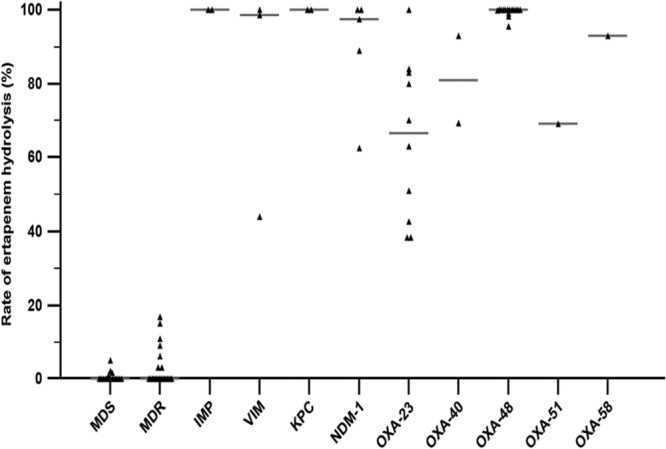
Rate of hydrolysis of ertapenem for strains susceptible to antibiotics (multidrug susceptible [MDS]), multidrug-resistant non-carbapenemase-producing strains (MDR), and carbapenemase-producing strains, including those producing IMP, VIM, KPC, NDM-1, OXA-23, OXA-40, and OXA-48, overproducing OXA-51, and producing OXA-58.
To our knowledge, this is the first report showing the use of UPLC-MS/MS in detecting carbapenemase activity in a broad range of bacterial species, including enterobacteria, P. aeruginosa, and A. baumannii. Like the phenotypic assays already described (3), the UPLC-MS/MS method requires overnight subculture of the strain and is able to detect only phenotypic expression of carbapenemase but no other resistance mechanisms (e.g., MDR pumps, loss of outer membrane proteins [OMP], etc.) that might also impair the clinical efficacy of certain carbapenems. Unlike molecular assays, the UPLC-MS/MS method did not allow identification of the carbapenemase enzyme, failing to provide molecular epidemiological information. Although molecular assays performed directly on the sample are faster than the UPLC-MS/MS method, the latter could be realized only by using an overnight subculture with a significant increased rapidity and sensitivity compared to MHT. Finally, despite the fact that the UPLC-MS/MS platform is clearly more costly than real-time PCR, detection of carbapenemase activity by UPLC-MS/MS could be easily implemented in laboratories already using this technology for measuring carbapenem antibiotic levels in patients. In conclusion, the UPLC-MS/MS assay described in this study is a fast and reliable method to detect carbapenemase activity for a strain irrespective of the enzyme type.
ACKNOWLEDGMENTS
We are indebted to Patrice Nordmann and Patrick Plésiat and all the staff of the French Reference Centre on Emerging Antibiotic Resistance for molecular characterization and determination of clone resistance.
We also thank Véronique Monteiller and Dominique Fayolle for their skillful technical assistance.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 2 December 2013
REFERENCES
- 1.Marchaim D, Navon-Venezia S, Schwaber MJ, Carmli Y. 2008. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbanemase, molecular characterization, epidemiology, and outcomes. Antimicrob. Agents Chemother. 52:1413–1418. 10.1128/AAC.01103-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Poirel L. 2013. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 68:487–489. 10.1093/jac/dks426 [DOI] [PubMed] [Google Scholar]
- 3.Wang P, Chen S, Guo Y, Xiong Z, Hu F, Zhu D, Zhang Y. 2011. Occurrence of false positive results for the detection of carbapenemases in carbapenemase-negative Escherichia coli and Klebsiella pneumoniae isolates. PLoS One 6:e26356. 10.1371/journal.pone.0026356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49:3321–3324. 10.1128/JCM.00287-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2441–2443. 10.1128/JCM.01002-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalhaes CG, Cayô R, Assis DM, Martins ER, Juliano L, Juliano MA, Gales AC. 2013. Detection of SPM-1-producing Pseudomonas aeruginosa and class D β-lactamase-producing Acinetobacter baumannii isolates by use of liquid chromatography-mass spectrometry and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51:287–290. 10.1128/JCM.02365-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. Twenty-first informational supplement. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 11.Denis C, Poirel L, Carricajo A, Grattard F, Fascia P, Verhoeven P, Gay P, Nuti C, Nordmann P, Pozzetto B, Berthelot P. 2012. Nosocomial transmission of NDM-1-producing Escherichia coli within a non-endemic area in France. Clin. Microbiol. Infect. 18:E128–130. 10.1111/j.1469-0691.2012.03761.x [DOI] [PubMed] [Google Scholar]
- 12.Poirel L, Ros A, Carrër A, Fortineau N, Carricajo A, Berthelot P, Nordmann P. 2011. Cross-border transmission of OXA-48-producing Enterobacter cloacae from Morocco to France. J. Antimicrob. Chemother. 66:1181–1182. 10.1093/jac/dkr023 [DOI] [PubMed] [Google Scholar]
- 13.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 14.Bonnin RA, Naas T, Poirel L, Nordmann P. 2012. Phenotypic, biochemical, and molecular techniques for detection of metallo-β-lactamase NDM in Acinetobacter baumannii. J. Clin. Microbiol. 50:1419–1421. 10.1128/JCM.06276-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedesch S, Cauda R, Bassetti M. 2012. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin. Infect. Dis. 55:943–950. 10.1093/cid/cis588 [DOI] [PubMed] [Google Scholar]



