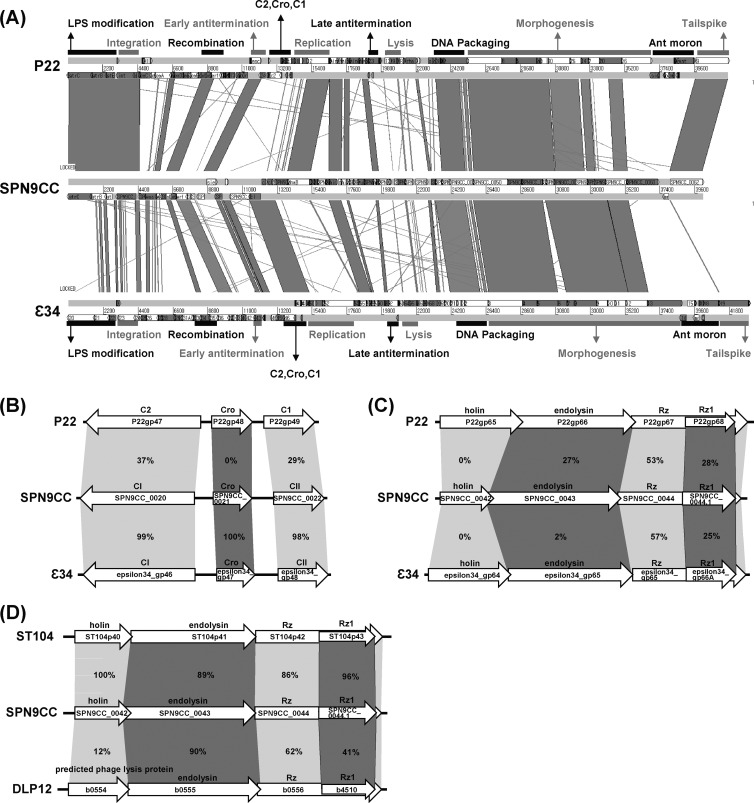
FIG 3.
Comparative genomic analysis of P22-like phages (SPN9CC, P22, ST104, and ε34) and E. coli K-12 prophage DLP12. (A) Comparative analysis of the complete genome sequences of SPN9CC, P22, and ε34 with BLASTN and ACT12. Black and gray bars indicate the functions of gene clusters in the genomes. (B and C) Comparative analyses of lysogeny control regions (B) and host cell lysis gene clusters (C) in SPN9CC, P22, and ε34. (D) Comparative analysis of host cell lysis gene clusters in SPN9CC, ST104, and E. coli K-12 prophage DLP12. The percentages of amino acid identity between homologous genes are indicated.