Abstract
Nine marine methanogenic Methanococcoides strains, including the type strains of Methanococcoides methylutens, M. burtonii, and M. alaskense, were tested for the utilization of N-methylated glycines. Three strains (NM1, PM2, and MKM1) used glycine betaine (N,N,N-trimethylglycine) as a substrate for methanogenesis, partially demethylating it to N,N-dimethylglycine, whereas none of the strains used N,N-dimethylglycine or sarcosine (N-methylglycine). Growth rates and growth yields per mole of substrate with glycine betaine (3.96 g [dry weight] per mol) were similar to those with trimethylamine (4.11 g [dry weight] per mol). However, as glycine betaine is only partially demethylated, the yield per methyl group was significantly higher than with trimethylamine. If glycine betaine and trimethylamine are provided together, trimethylamine is demethylated to dimethyl- and methylamine with limited glycine betaine utilization. After trimethylamine is depleted, dimethylamine and glycine betaine are consumed rapidly, before methylamine. Glycine betaine extends the range of substrates that can be directly utilized by some methanogens, allowing them to gain energy from the substrate without the need for syntrophic partners.
INTRODUCTION
Glycine betaine (N,N,N-trimethylglycine) is one of the most common compatible solutes in nature and is found in all three domains of life (1–3). In addition to its role in osmoadaptation, glycine betaine has been suggested to play a role in microbial cryoprotection and barotolerance (4, 5). Considering that intracellular glycine betaine concentrations can be some hundreds of millimoles per liter, depending on the salinity of the medium (6), it is clear that it must be very abundant in saline environments. For example, in hypersaline mats, total glycine betaine contents of up to 0.1 mmol per gram (dry weight) of sediment have been found (7).
In anoxic sediments, the addition of glycine betaine leads to methanogenic activity, but also to a simultaneous stimulation of sulfate reduction (8). However, the transient formation of similar amounts of trimethylamine (TMA) and acetate indicates that the reduction of betaine, as found in members of the genera Clostridium and Halanaerobacter (9, 10), is the first step in degradation. While acetate is utilized mainly by sulfate reducers, trimethylamine is a well-known noncompetitive substrate for methanogens (8, 11), allowing them to thrive within the sulfate reduction zone. This degradation pattern involving three different metabolic groups is quite complex, and it could be argued that it would be advantageous for the methanogens if they could demethylate glycine betaine directly, similar to direct choline (N,N,N-trimethylethanolamine) utilization, which has recently been documented (12). Although a number of methanogens have been tested, no glycine betaine consumption by methanogens has been reported so far (13–15).
In the present study, we demonstrate the partial demethylation of glycine betaine (N,N,N-trimethylglycine) to N,N-dimethylglycine (DMG) by members of the genus Methanococcoides. The potential implications of this novel methanogenic pathway are discussed.
MATERIALS AND METHODS
Sources of organisms.
In total, nine Methanococcoides strains were investigated. They included the three type strains Methanococcoides methylutens DSM 2657T, M. burtonii DSM 6242T, and M. alaskense DSM 17273T, obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) (Braunschweig, Germany), and five new Methanococcoides strains (AM1, DM1, NM1, PM1, and PM2) obtained from a range of marine habitats (12). Their 16S rRNA genes (GenBank numbers HE862406 to HE862410) share >99% similarity with that of M. methylutens DSM 2657T. One additional strain, MKM1, was isolated from an enrichment inoculated with sediment from the Meknes mud volcano of the Gulf of Cadiz with methylamine (MMA) as the substrate, using agar shake tubes (16). All cultures were incubated at 25°C.
Cultivation and media.
A bicarbonate-buffered and FeS-reduced artificial seawater medium (12, 17) was used for isolation, strain maintenance, and physiological experiments. The pH of the reduced medium was adjusted to 7.2 to 7.4 with sterile HCl or Na2CO3 if necessary. For enrichment and isolation, 10 mmol methylamine per liter was added.
For growth experiments, 150-ml serum bottles filled with 30 ml medium under an N2-CO2 (80/20 [vol/vol]) headspace and with 5 mmol of substrate per liter were used. Growth was monitored by the increase in headspace methane, and the specific growth rate (μ) was calculated from plots of the total accumulated methane against time (12, 18, 19). The growth yield was estimated from the increase in protein content analyzed by the method of Bradford (20).
Analytical techniques.
Headspace gas was measured by gas chromatography (PerkinElmer/Arnel Clarus [Sheldon, CT] 500 Natural Gas Analyzer), and the methane contents in the headspace and medium were calculated as described previously (12). Anions (including the organic acids acetate, lactate, and formate) were analyzed on a Dionex ICS-2000 Ion Chromatography System equipped with an AS50 autosampler (Dionex, Camberley, United Kingdom) (21).
Prior to ion chromatographic analysis, 1 ml of culture was centrifuged (15 min at 16,000 × g at 10°C), and the supernatant was diluted (1:10 [vol/vol]) in ultrapure water (>18.2 MΩ; Milli-Q system; Millipore). Cations (including ammonium, methylamines, betaine, and dimethylglycine) were analyzed using ion chromatography with nonsuppressed conductivity detection (22) on a Dionex ICS-2000 Ion Chromatograph equipped with a DS6 heated conductivity cell (45°C) and an AS50 autosampler (Dionex, Camberley, United Kingdom). Chromatographic separation was conducted on an Ionpac CS16 column at 50°C using methanesulfonic acid eluent (3 mmol · liter−1) and acetonitrile (10%) at a flow rate of 1.30 ml min−1.
RESULTS
Utilization of N-methylated glycines by Methanococcoides spp.
All Methanococcoides strains tested grew well with mono-, di-, and trimethylamine, and fresh methylamine-grown cultures were used to inoculate media with glycine betaine (N,N,N-trimethylglycine), DMG, or N-monomethylglycine (sarcosine) as the substrate. While none of the strains formed methane from DMG or sarcosine, three strains (NM1, PM2, and MKM1) produced methane from glycine betaine within 1 to 2 weeks. These positive results were confirmed by subcultivation on the same substrate. Negative cultures were incubated for at least 3 months, and regularly measured for methane production, since methanogenic cultures sometimes show very long lag phases (12).
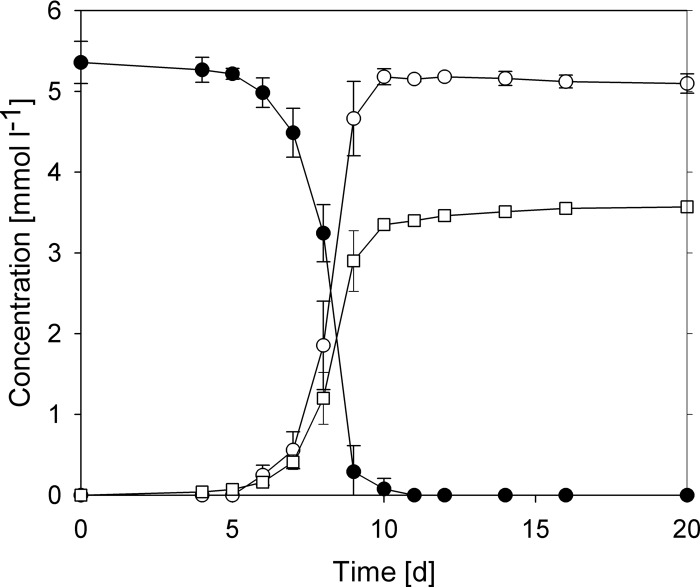
When the three strains were grown with glycine betaine, only a relatively small amount of methane was formed, with a methane-to-glycine betaine ratio of around 0.7. This suggested that glycine betaine was only partly demethylated. Since the three strains showed similar lag phases and growth rates, only one strain, NM1, was investigated in more detail. Ion chromatographic analysis identified DMG as the end product of methanogenesis from glycine betaine by strain NM1 (Fig. 1). At the end of the growth experiment, residual betaine concentrations were below the detection limit (130 μmol liter−1). After glycine betaine was consumed, the cultures were further incubated for a number of weeks but showed no decrease in DMG concentrations.
FIG 1.
Metabolism of glycine betaine by Methanococcoides sp. NM1. All values are the averages of three replicates, with the error bars indicating 1 standard deviation. □, methane; ●, glycine betaine; ○, N,N-dimethylglycine; d, days.
The maximum growth rate of strain NM1 with glycine betaine was 0.93 ± 0.01 day−1 (n = 3). This is a growth rate comparable to that of cultures with methylamine (0.96 day−1) but slightly lower than that of cultures with dimethylamine (DMA) (1.05 day−1) or trimethylamine (1.24 day−1) and higher than with methanol (0.64 day−1). On average, 0.97 mol of DMG and 0.67 mol of methane were formed per mole of betaine. The amount of protein formed in cultures with trimethylamine and glycine betaine was similar. However, as glycine betaine is only partially demethylated, the growth yield per methyl group is 3.96 g (dry weight) mol (methyl group)−1 and significantly higher than with mono-, di-, or trimethylamine (Table 1). Acetate, formate, and other organic acids were found only at minor concentrations (<0.04 mmol · liter−1).
TABLE 1.
Metabolic products and growth yields of Methanococcoides sp. strain NM1 grown on methylamine, dimethylamine, trimethylamine, and glycine betainea
| Substrate | Substrate consumed (mM) | Product formed (mM) |
Protein formed (mg liter−1) | Growth yield (g [dry wt] mol methyl group−1) | ||
|---|---|---|---|---|---|---|
| Ammonium | DMG | Methane | ||||
| Methylamine | 5.4 | 5.4 | 3.1 | 5.81 | 2.15 | |
| Dimethylamine | 5.1 | 5.1 | 6.7 | 8.95 | 1.75 | |
| Trimethylamine | 4.9 | 4.9 | 10.1 | 10.1 | 1.37 | |
| Betaine | 5.4 | 5.2 | 3.6 | 10.7 | 3.96 | |
All data are averages of triplicate cultures. The protein formed was converted into dry mass assuming that protein represents 50% of the dry weight (34).
Impact of trimethylamine on methanogenesis from glycine betaine by strain NM1.
Cultures of strain NM1 with trimethylamine and glycine betaine showed no clear diauxic substrate utilization (Fig. 2). As in previous studies (12, 13), TMA was first partially demethylated to DMA and MMA. However, although TMA was utilized first, there was some simultaneous decrease in glycine betaine in the presence of TMA. The highest rate of glycine betaine consumption occurred immediately after TMA was depleted, and this was simultaneous with DMA consumption. Strain NM1 utilized MMA only when glycine betaine and DMA were almost depleted. This pattern differs significantly from that found for Methanococcoides sp. strain AM1 in the presence of choline and TMA, where a significant lag occurred between the consumption of TMA and its intermediates and the start of choline utilization (12).
FIG 2.
Successive metabolism of trimethylamine, its intermediates, and glycine betaine by Methanococcoides sp. NM1. Both substrates were present in the medium from day 0. Note the different scale in the bottom graph showing the concentrations of intermediates of trimethylamine consumption. Only the first 10 days of the experiment are shown. The cultures were monitored for another 3 weeks but did not show any significant concentration changes. All values are the averages of three replicates, with the error bars indicating 1 standard deviation. □, methane; ●, glycine betaine; ○, N,N-dimethylglycine;
 , trimethylamine; ▼, ammonium; ◆ dimethylamine; ◊, methylamine.
, trimethylamine; ▼, ammonium; ◆ dimethylamine; ◊, methylamine.
Glycine betaine content in cells of Methanococcoides sp. strain NM1.
At the end of the growth experiment shown in Fig. 1, 1.5 ml of culture was washed in artificial seawater, and the cell pellet was resuspended in 1.5 ml of deionized water to lyse the cells. Cation analysis of three parallel cultures revealed the presence of N,N-dimethylglycine (353 ± 140 μmol · liter−1), Na+ (34 ± 10 mmol · liter−1), and K+ (0.69 ± 0.29 mmol · liter−1), but no glycine betaine, methylamines, or ammonium, in the cell pellets. In contrast, cells grown with trimethylamine (10 mmol · liter−1) contained significant concentrations of ammonium (53 μmol · liter−1), MMA (294 μmol · liter−1), DMA (41 μmol · liter−1), Na+ (5.3 mmol liter−1), and K+ (5.3 mmol liter−1), but no detectable glycine betaine or DMG.
DISCUSSION
Glycine betaine, a new substrate for methanogenic pure cultures.
In this study, we have shown the direct use of glycine betaine by pure cultures of methanogens. Previously, methanogenic degradation of glycine betaine was thought to require syntrophic interaction with a fermenter (or sulfate reducer) producing trimethylamine, which was then used by the methanogen (8, 13). However, like choline and N,N-dimethylethanolamine, which have recently been reported to be novel direct substrates for methanogens (12), glycine betaine can also be directly demethylated by methanogens. The presence of a syntrophic partner in our cultures can be ruled out, as no intermediates, TMA or acetate, were detected, which would have accumulated if glycine betaine was degraded by coculture.
At present, we can only speculate about how widespread the capacity to use glycine betaine is among methanogens. Like choline and N,N-dimethylethanolamine, glycine betaine is an N-methylated amine bearing a C2 side chain and belongs to a group of compounds that was thought not to support the growth of methanogenic pure cultures. Therefore, only a limited number of pure cultures belonging to the genera Methanococcoides, Methanosarcina, Methanohalophilus, and Methanomicrococcus (13–15, 23, 24) have been tested with glycine betaine or choline. However, choline and glycine betaine are not the only C2 methylated amines utilized by methanogens. Methanosarcina barkeri was shown to grow with N-ethyldimethylamine, but not with choline, glycine betaine, or N,N-diethylmethylamine (13). However, since N-ethyldimethylamine was considered of little biological significance, later studies neglected this substrate. Glycine betaine, in contrast, is a common osmolyte in saline environments (1, 3), and choline and N,N-dimethylethanolamine are headgroups of phospholipids present in anoxic sediments (25). Considering that three of the nine strains tested used glycine betaine and 5 out of 15 Methanococcoides spp. have been recently shown to utilize choline or N,N-dimethylethanolamine (12), it is clear that methanogens are more versatile than previously thought. Therefore, this physiological diversity, particularly with respect to N-methylated amines bearing a larger side chain, has been largely overlooked.
Whether glycine betaine is a direct substrate for methanogens in the marine environment needs to be investigated, although it is unlikely that they can compete with sulfate reducers for the substrate. Several sulfate reducers can utilize glycine betaine as an electron donor (26, 27), and it was shown that in intertidal sediments, sulfate reduction was strongly stimulated by the addition of glycine betaine (8). In sulfate-free layers, however, being able to use glycine betaine directly would make the methanogens independent of syntrophic interaction with fermenters, some of which may not release trimethylamine that could then be used by the methanogens and therefore would restrict methanogenesis. For example, in the presence of glycine betaine when methanogens were inhibited in intertidal sediments by the addition of 2-bromoethanesulfonate (BES), less than 60% of the theoretically possible TMA was formed (8). This indicates that either not all of the betaine is degraded via trimethylamine or that some of the TMA is used by other processes, such as homoacetogenesis.
Incomplete degradation of glycine betaine.
All three strains utilizing glycine betaine only partially demethylated their substrates to N,N-dimethylglycine. This may be surprising, particularly considering that the Methanococcoides spp. using choline demethylated their substrates completely to ethanolamine (12). However, a range of organisms also produce DMG from glycine betaine, including several Desulfobacterium spp. and Acetobacterium spp. (26, 28). In addition, Eubacterium limosum converts glycine betaine and CO2 into DMG, acetate, and butyrate (29), while some homoacetogens, like Sporomusa spp., ferment glycine betaine into acetate, trimethylamine, and DMG (30).
The demethylation of glycine betaine to DMG or glycine produces −180.4 and −248.2 kJ per mol of glycine betaine, respectively (Table 2). This means that the first methyl group yields more than five times more energy than the other two. This high energy yield may also explain the relatively high growth yield observed for growth on glycine betaine (Table 1). However, the ΔG°′ for the demethylation of DMG to glycine is still −67.8 kJ per mol DMG, and considering that DMG has two methyl groups, the ΔG°′ per methyl group is comparable to the value for methylamine (−43.0 kJ per mol). However, although it seems a potential waste of energy, the cultures investigated here did not utilize the DMG produced even after prolonged incubation of several weeks.
TABLE 2.
Equations and free energies of reaction for the methanogenic degradation of glycine betaine to N,N-dimethylglycine (equation 1), glycine betaine to glycine (equation 2), N,N-dimethylglycine to glycine (equation 3), sarcosine to glycine (equation 4), and methanogenesis from methylamine (equation 5)
| Equation no. | Reaction | ΔG°′a (kJ/reaction) |
|---|---|---|
| 1 | 4 (CH3)3N+CH2COO− + 2 H2O→4 (CH3)2NH+CH2COO− + 3 CH4 + CO2 | −721.7 |
| 2 | 4 (CH3)3N+CH2COO− + 6 H2O→4 H3N+CH2COO− + 9 CH4 + 3 CO2 | −992.8 |
| 3 | 2 (CH3)2NH+CH2COO− + 2 H2O→2 H3N+CH2COO− + 3 CH4 + CO2 | −135.5 |
| 4 | 4 (CH3)NH2+CH2COO− + 2 H2O→4 H3N+CH2COO− + 3 CH4 + CO2 | −157.7 |
| 5 | 4 (CH3)NH3+ + 2 H2O→4 NH4+ + 3 CH4 + CO2 | −172.1 |
ΔGf°′ values for the single compounds were taken from Jankowski et al. (35, supplemental material). ΔGf°′ values for glycine betaine (−129.8 kJ mol−1), N,N-dimethylglycine (−306.6 kJ mol−1), and sarcosine (−331.3 kJ mol−1) were estimated using the group contribution method described by Jankowski et al. (35). All values are calculated for standard conditions (298 K; pH 7; 1 atm) in aqueous systems and for the predominant ions at neutral pH.
Glycine betaine as a compatible solute in Methanococcoides sp. NM1.
Both glycine betaine and DMG have been documented as compatible solutes in halotolerant and halophilic methanogenic archaea (31–33). However, cells of strain NM1 grown in artificial seawater with trimethylamine as the substrate did not contain any detectable amounts of glycine betaine but showed a slight accumulation of K+ plus significant amounts of methylamine. This is similar to other methanogens, like Methanosarcina spp., that can accumulate K+ for osmoregulation and synthesize the amino acids α-glutamate and Nε-acetyl-β-lysine as osmolytes but can take up glycine betaine if it is present in the medium (33). However, the uptake and accumulation of glycine betaine in Methanosarcina spp. suppresses the formation of other osmolytes, which is thought to save significant energy. Cells of strain NM1 might not only save energy by taking up glycine betaine instead of synthesizing other osmolytes, they also can use glycine betaine as a metabolic substrate. Since DMG acts as a compatible solute, as well, this means that the partial demethylation of glycine betaine allows energy generation and energy saving by the metabolic end product being an osmoregulant.
ACKNOWLEDGMENTS
This research was funded by the Natural Environmental Research Council, United Kingdom (NE/F00477X/1 and NE/F018983/1), and the European Community's Seventh Framework Programme (FP7/2007–2013) under the HERMIONE project, grant agreement no. 226354.
We thank Laurent Toffin for providing sediment samples from the Napoli mud volcano, from which strain NM1 was isolated; Bettina Buchmann for helping with protein analysis; Detlef Jensen (DIONEX) for his expertise; and three anonymous reviewers for their support.
Footnotes
Published ahead of print 25 October 2013
REFERENCES
- 1.Galinski EA. 1995. Osmoadaptation in bacteria. Adv. Microb. Physiol. 37:273–328. 10.1016/S0065-2911(08)60148-4 [DOI] [PubMed] [Google Scholar]
- 2.Roeßler M, Müller V. 2001. Osmoadaptation in bacteria and archaea: common principles and differences. Environ. Microbiol. 3:743–754. 10.1046/j.1462-2920.2001.00252.x [DOI] [PubMed] [Google Scholar]
- 3.Ventosa A, Nieto JJ, Oren A. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casanueva A, Tuffin M, Craig C, Cowan DA. 2010. Molecular adaptations to psychrophily: the impact of ‘omic' technologies. Trends Microbiol. 18:374–381. 10.1016/j.tim.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 5.Smiddy M, Sleator RD, Patterson MF, Hill C, Kelly AL. 2004. Role for compatible solutes glycine betaine and L-carnithine in Listerial barotolerance. Appl. Environ. Microbiol. 70:7555–7557. 10.1128/AEM.70.12.7555-7557.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imhoff JH, Rodriguez-Valera F. 1984. Betaine is the main compatible solute of halophilic eubacteria. J. Bacteriol. 160:478–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King GM. 1988. Methanogenesis from methylated amines in a hypersaline algal mat Appl. Environ. Microbiol. 54:130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King GM. 1984. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl. Environ. Microbiol. 48:719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouné S, Manac'h N, Hirschler A, Caumette P, Willison JC, Matheron R. 1999. Haloanaerobacter salinarius sp. nov., a novel halophilic fermentative bacterium that reduces glycine-betaine to trimethylamine with hydrogen or serine as electron donors; emendation of the genus Haloanaerobacter. Int. J. Syst. Bacteriol. 49:103–112. 10.1099/00207713-49-1-103 [DOI] [PubMed] [Google Scholar]
- 10.Naumann E, Hippe H, Gottschalk G. 1983. Betaine: new oxidant in the Stickland reaction and methanogenesis from betaine and L-alanine by a Clostridium sporogenes-Methanosarcina barkeri coculture. Appl. Environ. Microbiol. 45:474–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oremland RS, Marsh LM, Polcin S. 1982. Methane production and simultaneous sulfate reduction in anoxic, salt marsh sediments. Nature 296:143–145. 10.1038/296143a0 [DOI] [Google Scholar]
- 12.Watkins AJ, Roussel EG, Webster G, Parkes RJ, Sass H. 2012. Choline and N,N-dimethylethanolamine as direct substrates for methanogens. Appl. Environ. Microbiol. 78:8298–8303. 10.1128/AEM.01941-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hippe H, Caspari D, Fiebig K, Gottschalk G. 1979. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation in Methanosarcina barkeri. Proc. Natl. Acad. Sci. U. S. A. 76:494–498. 10.1073/pnas.76.1.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprenger WW, van Belzen MC, Rosenberg J, Hackstein JHP, Keltjens JT. 2000. Methanomicrococcus blatticola gen. nov., sp nov., a methanol- and methylamine-reducing methanogen from the hindgut of the cockroach Periplaneta americana. Int. J. Syst. Evol. Microbiol. 50:1989–1999. 10.1099/00207713-50-6-1989 [DOI] [PubMed] [Google Scholar]
- 15.Tanaka K. 1994. Anaerobic degradation of tetramethylammonium by a newly isolated marine methanogen. J. Ferment. Bioeng. 78:386–388. 10.1016/0922-338X(94)90287-9 [DOI] [Google Scholar]
- 16.Parkes RJ, Sass H, Webster G, Watkins AJ, Weightman AJ, O'Sullivan LA, Cragg BA. 2010. Methods for studying methanogens and methanogenesis in marine sediments, p 3799–3827 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology, vol 5 Springer, Berlin, Germany [Google Scholar]
- 17.Süß J, Engelen B, Cypionka H, Sass H. 2004. Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol. Ecol. 51:109–121. 10.1016/j.femsec.2004.07.010 [DOI] [PubMed] [Google Scholar]
- 18.Lyimo TJ, Pol A, Jetten MSM, op den Camp HJM. 2009. Diversity of methanogenic archaea in a mangrove sediment and isolation of a new Methanococcoides strain. FEMS Microbiol. Lett. 291:247–253. 10.1111/j.1574-6968.2008.01464.x [DOI] [PubMed] [Google Scholar]
- 19.Powell GE. 1983. Interpreting gas kinetics of batch cultures. Biotechnol. Lett. 5:437–440. 10.1007/BF00132224 [DOI] [Google Scholar]
- 20.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 21.Webster G, Blazejak A, Cragg BA, Schippers A, Sass H, Rinna J, Tang X, Mathes F, Ferdelman T, Fry JC, Weightman AJ, Parkes RJ. 2009. Subsurface microbiology and biogeochemistry of a deep, cold-water carbonate mound from the Porcupine Seabight (IODP Expedition 307). Environ. Microbiol. 11:239–257. 10.1111/j.1462-2920.2008.01759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang JJ, Zhu Y. 2007. Determination of betaine, choline and trimethylamine in feed additive by ion-exchange liquid chromatography/non-suppressed conductivity detection. J. Chromatogr. A 1170:114–117. 10.1016/j.chroma.2007.09.014 [DOI] [PubMed] [Google Scholar]
- 23.Heijthuijsen JHFG, Hansen TA. 1989. Betaine fermentation and oxidation by marine Desulfuromonas strains. Appl. Environ. Microbiol. 55:965–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paterek JR, Smith PH. 1988. Methanohalophilus mahii gen. nov., sp. nov., a methylotrophic halophilic methanogen. Int. J. Syst. Bacteriol. 38:122–123. 10.1099/00207713-38-1-122 [DOI] [PubMed] [Google Scholar]
- 25.Seidel M, Graue J, Engelen B, Köster J, Sass H, Rullkötter J. 2012. Advection and diffusion determine the vertical distribution of microbial communities in intertidal sediments as revealed by combined biogeochemical and molecular biological analysis. Org. Geochem. 52:114–129. 10.1016/j.orggeochem.2012.08.015 [DOI] [Google Scholar]
- 26.Heijthuijsen JHFG, Hansen TA. 1989. Anaerobic degradation of betaine by marine Desulfobacterium strains. Arch. Microbiol. 152:393–396. 10.1007/BF00425179 [DOI] [Google Scholar]
- 27.Schink B, Thiemann V, Laue H, Friedrich MW. 2002. Desulfotignum phosphitoxidans sp. nov., a new marine sulfate reducer that oxidizes phosphite to phosphate. Arch. Microbiol. 177:381–391. 10.1007/s00203-002-0402-x [DOI] [PubMed] [Google Scholar]
- 28.Van der Maarel MJEC, Jansen M, Haanstra R, Meijer WG, Hansen TA. 1996. Demethylation of dimethylsulfoniopropionate to 3-S-methylmercaptopropionate by marine sulfate-reducing bacteria. Appl. Environ. Microbiol. 62:3978–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller E, Fahlbusch K, Walther R, Gottschalk G. 1981. Formation of N,N-dimethylglycine, acetic acid, and butyric acid from betaine by Eubacterium limosum. Appl. Environ. Microbiol. 42:439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Möller B, Oßmer R, Howard BH, Gottschalk G, Hippe H. 1984. Sporomusa, a new genus of Gram-negative anaerobic bacteria including Sporomusa sphaeroides spec. nov. and Sporomusa ovata spec. nov. Arch. Microbiol. 139:388–396. 10.1007/BF00408385 [DOI] [Google Scholar]
- 31.Menaia JAGF, Duarte JC, Boone DR. 1993. Osmotic adaptation of moderately halophilic methanogenic archaeobacteria, and detection of cytosolic N,N-dimethylglycine. Experientia 49:1047–1054. 10.1007/BF01929912 [DOI] [Google Scholar]
- 32.Robertson DE, Noll D, Roberts MF, Menaia JAGF, Boone DR. 1990. Detection of the osmoregulator betaine in methanogens. Appl. Environ. Microbiol. 56:563–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sowers KR, Gunsalus RP. 1995. Halotolerance in Methanosarcina spp.: role of Nε-acetyl-β-lysine, α-glutamate, glycine betaine and K+ as compatible solutes for osmotic adaptation. Appl. Environ. Microbiol. 61:4382–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Archer DB. 1984. Detection and quantitation of methanogens by enzyme-linked immunosorbent assay. Appl. Environ. Microbiol. 48:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. 2008. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 95:1487–1499. 10.1529/biophysj.107.124784 [DOI] [PMC free article] [PubMed] [Google Scholar]