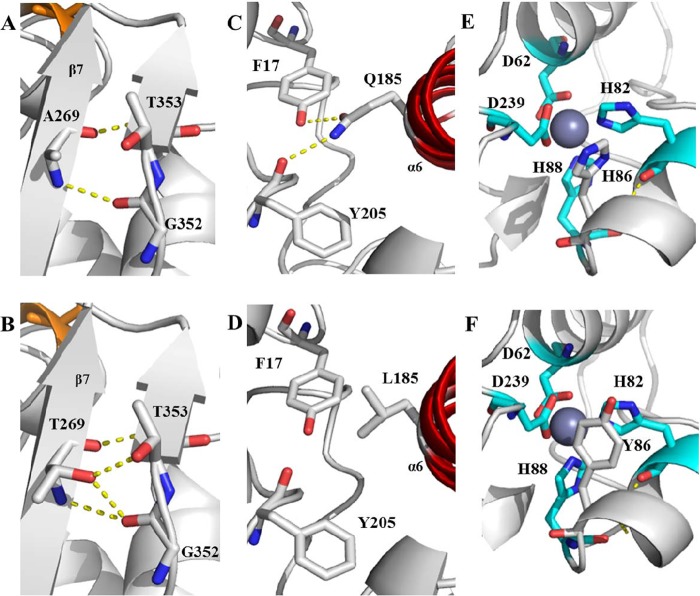
FIG 7.
Structure models with hydrogen bonds for wild-type lipase T6 and the improved mutants. (A) Wild-type A269; (B) A269T; (C) wild-type Q185; (D) Q185L; (E) wild-type H86; (F) H86Y. The α-helix lid (α6) is marked in red (C and D), and the zinc-binding residues are marked in cyan, with zinc in dark gray. Possible hydrogen bonds are marked by yellow dashed lines. The models were visualized using PyMOL based on the crystal sturucture of G. stearothermophilus P1 lipase (PDB code 1JI3).