Abstract
Dental caries induced by Streptococcus mutans is one of the most prevalent chronic infectious diseases worldwide. The pathogenicity of S. mutans relies on the bacterium's ability to colonize tooth surfaces and survive a strongly acidic environment. We performed an ISS1 transposon mutagenesis to screen for acid-sensitive mutants of S. mutans and identified an SMU.746-SMU.747 gene cluster that is needed for aciduricity. SMU.746 and SMU.747 appear to be organized in an operon and encode a putative membrane-associated permease. SMU.746- and SMU.747-deficient mutants showed a reduced ability to grow in acidified medium. However, the short-term or long-term acid survival capacity and F1F0 ATPase activity remained unaffected in the mutants. Furthermore, deletion of both genes did not change cell membrane permeability and the oxidative and heat stress responses. Growth was severely affected even with slight acidification of the defined medium (pH 6.5). The ability of the mutant strain to acidify the defined medium during growth in the presence of glucose and sucrose was significantly reduced, although the glycolysis rate was only slightly affected. Surprisingly, deletion of the SMU.746-SMU.747 genes triggered increased biofilm formation in low-pH medium. The observed effects were more striking in a chemically defined medium. We speculate that the SMU.746-SMU.747 complex is responsible for amino acid transport, and we discuss its possible role in colonization and survival in the oral environment.
INTRODUCTION
Streptococcus mutans is a principal etiological agent of dental caries (1, 2). Among all the physiological traits, three play crucial roles in S. mutans pathogenicity. First, the ability to form biofilm allows bacteria to attach to and colonize the tooth surface. There are two major mechanisms that dictate initial attachment and biofilm formation by S. mutans: sucrose-dependent and sucrose-independent mechanisms (3). A crucial role in sucrose-dependent adherence is modulated by two extracellular enzymes, glucosyltransferases (GTFs) and fructosyltransferases (FTFs), and by glucan-binding proteins (GBP) (4), whereas sucrose-independent adherence is controlled primarily by SpaP, a major surface antigen of S. mutans (5). The second trait is the ability of the bacterium to produce organic acids (acidogenesis) and reduce the pH of the environment to well below 4.0. Acidogenesis is an effect of postglycolysis processes that occur under anaerobic conditions, where pyruvate is fermented primarily to lactic acid. However, under a low-glucose condition, other organic acids, such as acetic acid, formic acid, and ethanol, are produced (6, 7).
The third important trait is the ability to survive in a low-pH environment (aciduricity). It is well known that this organism can grow and carry out glycolysis at pH 5.0 or lower and survive highly acidic conditions (2). There are a few mechanisms of aciduricity in S. mutans that have recently been studied to some extent (8, 9). The most important mechanism depends on the proton-extruding F1-F0 ATPase activity, which is induced and functions well at pH 5.0 and below, allowing the organism to maintain a proper pH gradient across the membrane (9, 10). Other mechanisms of acid resistance in S. mutans include induction of stress proteins (11, 12), changes in membrane-associated proteins and fatty acid composition (13), DNA repair enzymes (14, 15), and an increase in alkali production through several metabolic pathways (16, 17). Recently, it has been shown that the change of carbon flux from pyruvate production to branched-chain amino acid biosynthesis also helps to maintain internal pH (18). In addition to these, biofilm formation can be also responsible for acid resistance in S. mutans (19). Biofilm-embedded bacteria are in general more resistant to several environmental conditions than their planktonic counterparts (20, 21).
S. mutans UA159 contains more than 280 genes associated with various transport systems, accounting for almost 15% of the total open reading frames (ORFs) (22). All three types of transport mechanisms, passive or facilitated diffusion, energy-driven symporters, and antiporters, are present in S. mutans. Putative transport proteins have been identified for the uptake of essential inorganic ions, efflux of toxic metal ions, and undefined molecules (23). Similar transport systems have been identified for amino acids, carbohydrates, oligopeptides, osmoprotectants (proline/glycine betaine and choline), bacteriocins, and DNA (22). The fact that streptococci are equipped with a broad range of transporters that positively impact bacterial fitness and virulence (23–27) raises the proposition that these organisms prefer uptake, breakdown, and catabolism of nutrients, peptides, and amino acids instead of de novo synthesis.
In this study, using transposon mutagenesis, we identified two S. mutans UA159 genes, SMU.746 and SMU.747, that are involved in acidogenesis, biofilm formation, and low-pH survival. The SMU.747 and SMU.746 genes are organized in an operon and encode a two-component permease system. The data presented here indicate that the SMU.746-SMU.747 membrane permease system is responsible for a specific transport of amino acid residue. We have shown that this permease plays a very important role in all three major traits responsible for the pathogenicity of S. mutans.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The S. mutans strains and plasmids used in this study are listed in Table 1. Escherichia coli EC100 was grown in Luria-Bertani medium supplemented with 100 μg/ml ampicillin (Ap), 300 μg/ml erythromycin (Em), or 50 μg/ml kanamycin (Km) when necessary. S. mutans strains were routinely grown in Todd-Hewitt medium (BBL, BD) supplemented with 0.2% yeast extract (THY medium), TY medium (1.4% Tryptone and 0.8% yeast extract), or CDM minimal medium with 1% carbon source (28). For some experiments, amino acids in CDM were replaced with peptone (20 mg/ml). When necessary, 5 μg/ml erythromycin (Em) or 300 μg/ml kanamycin (Km) was added to THY medium, and the pH of the medium was adjusted with HCl or potassium phosphate and sodium citrate (50 mM). For measuring growth kinetics, overnight cultures of S. mutans UA159 and its derivative mutants were grown in THY and diluted ∼25-fold in fresh THY medium (buffered at a specific pH) to get an equal initial cell density (optical density at 595 nm [OD595], Klett Units). Growth kinetics were monitored at 37°C for 24 h using a Klett colorimeter, model 900-3 (Bel-Art Scienceware, New Jersey, USA).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| S. mutans | ||
| UA159 | Wild type, serotype c | 22 |
| IBS16 | UA159 derivative, ΔSMU.746, Kmr | This study |
| IBSL28 | UA159 derivative, ΔSMU.747, Kmr | This study |
| IBSL23 | UA159 derivative, ΔSMU.745, Kmr | This study |
| IBSL32 | UA159 derivative, ΔSMU.746-SMU.747, Kmr | This study |
| IBSL32ΔK | IBSL32 derivative, Kms | This study |
| 3S3F | UA159 with ISS1 inserted in codon 146 of SMU.747 | This study |
| 7A5G | UA159 with ISS1 inserted in codon 172 of SMU.747 | This study |
| 61B11 | UA159 with ISS1 inserted in codon 177 of SMU.747 | This study |
| E. coli | ||
| EC100 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galKλ− rpsL (strR) nupG | Epicentre |
| Plasmids | ||
| pGEM-T EZ | Commercial TA cloning vector, Apr | Promega |
| pUC18 | Commercial cloning vector, Apr | Invitrogen |
| pIB107 | GusA promoter probe vector, Kmr | 34 |
| pIB184-Km | S. mutans expression shuttle vector, Kmr | 33 |
| pIBC62 | pGEM-T EZ derivative with a 2.27-kb fragment containing SMU.747 and SMU.746, Apr | This study |
| pIBC77 | pGEM-T EZ derivative with a 1.34-kb fragment containing SMU.747, Apr | This study |
| pIBL09 | pUC18 derivative with a 1.73-kb fragment containing SMU.745, Apr | This study |
| pIBL17 | pIBL09 derivative for deletion of SMU.745, Apr Kmr | This study |
| pIBL20 | pIBC62 derivative for deletion of SMU.746, Apr Kmr | This study |
| pIBL24 | pIBC77 derivative for deletion of SMU.747, Apr Kmr | This study |
| pIBL28 | pIBC62 derivative for deletion of SMU.746, Apr Kmr | This study |
| pIBL30 | pIB107 with 373-bp SMU.747 promoter fragment, Kmr | This study |
| pIBL31 | pIB107 with 163-bp SMU.745 promoter fragment, Kmr | This study |
| pIBL36 | pIB184K expressing SMU.746-SMU.747, Apr Kmr | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Emr, erythromycin resistance.
Transposition assay, screening, and identification of ISS1 insertion sites.
S. mutans mutagenesis with ISS1 was performed as described earlier (23). Mutant cultures were spotted onto THY pH 5.5 and pH 7.0 agar plates with a 48-pin replicator (Sigma) and incubated at 37°C in CO2 incubator. Colonies that grew on THY plates at pH 7.0 but failed to grow on THY plates at pH 5.5 were cultured overnight in THY-Em at 37°C and processed for analysis. The location of the inserted ISS1 element was then identified by inverse PCR using the primers ISS1Rout1 and ISS1Fout1 as described previously (29). The PCR products were purified from agarose gel and sequenced with the primer ISS1-Rout2. The flanking sequences obtained from sequencing analysis were mapped on the genome of S. mutans UA159 by a BLAST search. Genome-wide synteny of the SMU.746-SMU.747 region was analyzed by using the software tool WebACT (30).
Construction of SMU.746, SMU.747, and SMU.745 deletion mutants.
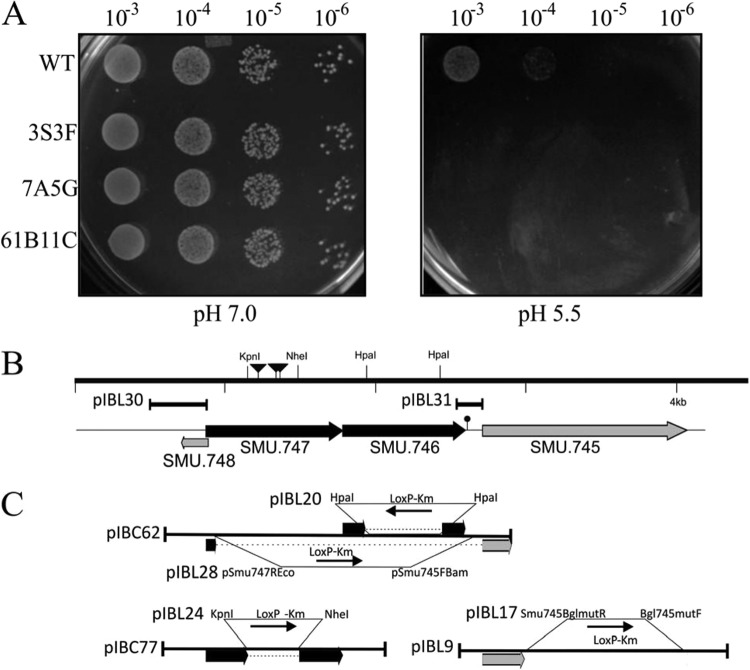
Construction scheme for the SMU.745, SMU.746, or SMU.747 single-gene deletion and SMU.746/SMU.747 double deletion mutants is shown on Fig. 1. Briefly, PCR fragments containing both SMU.747 and SMU.746 or containing only SMU.747 were cloned into pGemT-Easy to generate pIBC62 and pIBC77, respectively. Similarly, a PCR fragment containing the SMU745 gene was cloned into pUC18 to generate pIBL9. To construct SMU.746 and SMU.747 gene deletions, a 1.307-kb kanamycin resistance gene with flanking loxP sequences (LoxP-Kmr) was amplified from pIB-D38 (31) and cloned into blunted KpnI/NheI sites of the pIBC77 or HpaI site of the pIBC62 plasmid to create pIBL24 or pIBL20, respectively. To construct the SMU.745 mutant, a PCR fragment was first obtained using an inverted PCR (iPCR) with the primers BglSMU745mutR and BglSMU745mutF (see Table S1 in the supplemental material). The amplicon was then ligated with the LoxP-Kmr cassette, which resulted in the pIBL17 plasmid. A similar technique was used to obtain a double SMU.747-SMU.746 mutant. An iPCR fragment obtained with the pSMU.747Eco and pSmu745Bam primers was ligated with the LoxP-Kmr cassette, generating the pIBL28 plasmid. PCR fragments amplified with the universal M13/pUC primers were used for transformation of S. mutans UA159. Kanamycin-resistant transformants were isolated and analyzed by PCR to confirm the specific deletion. Successful representative transformants were chosen and named IBSL28 (ΔSMU.747), IBSL16 (ΔSMU.746), IBSL23 (ΔSMU.745), and IBSL32 (ΔSMU.747/SMU.746).
FIG 1.
(A) Confirmation of acid-sensitive phenotype. ISS1 transposon mutants that displayed an initial acid-sensitive phenotype were further verified by spotting of 5.0 μl from a 10-fold dilution series, with a starting optical density (A600) of 2.0 made in 0.85% NaCl, onto THY agar plates at pH 5.5 or 7.0. Experiments were repeated at least three times, and the relevant areas of the representative plates are shown. UA159 is the wild-type strain, while 3S3F, 7A5G, and 61B11C are independent mutants. (B) Genetic organization of the SMU.745-SMU.747 region of S. mutans UA159. Locations of ISS1 insertion are shown with inverted triangles. The lollipop indicates putative transcription termination. Plasmids used for promoter analysis are indicated. (C) Plasmids used for construction of the deletion mutants and complementation are shown. Arrows indicate gene orientation.
Construction of a complementing strain.
The plasmid pIBC62, containing both the SMU.747 and SMU.746 genes, was cloned into the BamHI site of the pIB184Km plasmid (32). Clones selected on Km (50 μg/ml) or Ap (100 μg/ml) were checked by PCR for presence and correct insert orientation. The correct construct, named pIBL36, was used for transformation of the IBSL32 strain.
β-Glucuronidase (Gus) assay.
PCR products containing the SMU.747 (373 bp; −370 to +3) and SMU.745 (163 bp; −160 to +3) promoters (Fig. 1) were cloned into the SmaI site of pIB107 (33), resulting in the pIBL30 and pIBL31 plasmids, respectively. S. mutans cultures containing these plasmids were grown overnight and were diluted 1:20 and grown in THY broth to exponential phase. For acid induction, 5 ml of culture was harvested, washed with saline, and resuspended in 5 ml of THY broth at pH 5.5 for 2 h, a signal pH that has been demonstrated to induce acid adaptation of S. mutans effectively (11); cells grown at pH 7.0 were used as a control. GusA activity was measured as described earlier (34).
General stress response.
Sensitivity of the S. mutans mutant strains to puromycin, hydrogen peroxide, and methyl viologen was evaluated by growth on THY agar plates containing these reagents as described previously (23).
Biofilm formation and analysis.
For biofilm formation on a polystyrene surface, flat-bottom 96-well microtiter plates (Corning Inc.) were used. S. mutans overnight cultures were diluted 1:40 in fresh medium, and 150-μl aliquots were dispensed into wells. After 24 h of incubation (37°C, 5% CO2), cell density was measured (OD595) using a Biotek plate reader, and 30 μl of Gram crystal violet (Remel) was applied for staining for 1 h. Plates were washed with water and air dried, and crystal violet was solubilized with an ethanol-acetone (4:1) solution. The OD570 was determined from this solution, and the biofilm amount was calculated as the ratio of OD570 to OD595.
For microscopy analysis, 1 ml of the 1:40 overnight culture dilutions were also used to inoculate wells of an four-well glass chamber slide (Lab-Tek; Nalge) for biofilm formation on a glass surface. After 24 h of incubation (37°C, 5% CO2), biofilms were stained for 1 h with 1.5 μM BacLight green (Molecular Probes) in 0.9% saline. Slides were analyzed with a standard fluorescent (Nikon Eclipse E600) or confocal laser scanning microscopy (CLSM) (Leica TCS-SPE).
Short-term acid killing and long-term acid survival.
The ability of cells to withstand acid challenge was determined as described previously (35). Briefly, S. mutans strains were grown in THY medium. Cultures were harvested (at an OD600 of ≅0.3) by centrifugation at 3,800 × g at 4°C for 10 min, washed once with 0.1 M glycine buffer, pH 7.0, and then subjected to killing by incubating the cells in 0.1 M glycine buffer, pH 2.8, for 0, 12.5, 25, 32.5, and 50 min. The surviving cells were appropriately diluted, plated on THY plates, and incubated in 5% CO2 at 37°C for 24 h.
Long-term acid survival was determined as described elsewhere (36). Briefly, an overnight culture was diluted 1:20 in TY or CDM medium with 1% glucose. Cultures were allowed to grow for 24 h, at which point aliquots were removed for serial dilution in 0.1 M glycine and plating on THY agar. Refreshed cultures continued to be incubated at 37°C in 5% CO2 for several days, with serial dilutions of the cultures plated daily until growth was no longer detected. Colonies were counted after 24 h of incubation.
Cell permeabilization assay.
Cell permeabilization was performed as described previously (37). Briefly, samples of 100 ml of cultures were centrifuged in the cold, and cells from each sample were resuspended in 5 ml of 75 mM Tris-HCl buffer (pH 7.0) with 10 mM MgSO4. Toluene (550 μl) was added to each cell suspension prior to vortex mixing and incubation for 5 min at 37°C. Each cell suspension was then subjected to two cycles of freezing in a dry ice-ethanol bath and thawing at 37°C. Permeabilized cells were harvested by centrifugation and resuspended in 2.5 ml of 75 mM Tris-HCl buffer (pH 7.0) with 10 mM MgSO4. Aliquots (125 μl) of suspension were quickly frozen in a dry ice-ethanol bath and stored at −70°C. The protein concentration was measured using the Quick Start Bradford method (Bio-Rad Laboratories, Inc., California) after trichloroacetic acid (TCA) precipitation.
ATPase assays.
An 125-μl sample of permeabilized cell suspension was added to 3.0 ml of 50 mM Tris-maleate buffer (pH 6.0) with 10 mM MgSO4, and the mixture was warmed to room temperature. The ATPase reaction was initiated by the addition of 30 μl of 0.5 M ATP (pH 6.0). Samples of 25 μl (each) were removed and assayed for inorganic phosphate liberated from cleavage of ATP by using a phosphate colorimetric assay kit (BioVision, Inc., California). ATPase activities were expressed as millimoles of phosphate released from ATP per microgram of protein.
Proton permeability assays.
Proton permeability assays were performed as described elsewhere (18). Briefly, 200-ml cultures of the wild-type (WT) and IBSL32 mutant strains were grown in THY medium at 37°C in 5% CO2 overnight. Cells were harvested by centrifugation, washed in 5 mM MgCl2, resuspended in 20 mM K3PO4 buffer, pH 7.2, 50 mM KCl, and 1 mM MgCl2, and incubated for 2 h. After incubation, cells were harvested and resuspended to a 20-mg/ml concentration. Five-milliliter aliquots were titrated to pH ∼4.7 by the addition of 10 mM HCl–50 mM KCl, and the pH values were recorded. At the 50-min time point, 10% (vol/vol) butanol was added to disrupt the cell membrane and allow the cytoplasmic and external pHs to equilibrate. At the 80-min time point, final pH values were recorded. The experiment was performed in triplicate using three independent cultures.
Terminal-pH assay.
Cultures of S. mutans UA159 and the mutant derivatives were grown in THY and TY supplemented with 1.0% glucose at the initial pH-unadjusted state and at pHs 7.0 and 5.5 at 37°C in an atmosphere of 5% CO2 for 18 h prior to terminal-pH measurement. The final pH in CDM medium with 1% glucose or sucrose was measured after 40 h of incubation at 37°C. The cell density of 150 μl of cultures per well in a 96-well plate was measured with a Biotek HT plate reader.
Glycolysis pH drop-down analysis.
S. mutans strains from THY overnight cultures were harvested, washed once with salt solution (50 mM KCl and 1 mM MgCl2), incubated for 40 min at 37°C, and resuspended in 0.05 M phosphate-citrate buffer at pH 7.0 or pH 5.0 (20 mg [wet mass]/ml). Cell mixtures were induced with 1% glucose, and acid presence was tested by pH measurement for 2 h.
RESULTS
Identification of genes involved in low-pH survival.
To isolate genes that are potentially involved in low-pH survival, we used ISS1 transposition mutagenesis in S. mutans strain UA159 as described previously (23, 29). Screening a collection of mutants grown on THY pH 5.5 agar plates allowed us to select nine mutants sensitive to low pH (Fig. 1A). Using an inverse PCR method, we were able to identify the ISS1 insertion sites in seven sensitive strains. Four of the insertion sites were located within or in the vicinity of the SMU.747 and SMU.746 genes (Fig. 1B), while three other insertions were in SMU.309, SMU.1690, and SMU.1781. SMU.1690 encodes an integral membrane protein involved in d-alanine export, and it was previously shown that this gene is involved in the acid tolerance response (38). Thus, the plate-based screening method that we used here is capable of identifying genes that are indeed responsible for aciduricity. Since we obtained multiple insertions in the SMU.746-SMU.747 locus, we focused on characterizing this locus further.
Characterization of the SMU.747-SMU.746 permease locus.
The genomic DNA fragment adjacent to the ISS1 insertion sites contains four described genes (Fig. 1). Three of those genes, SMU.747, SMU.746, and SMU.745, are transcribed in the same direction, while the fourth, a small (181-bp) gene (SMU.748), is transcribed in the opposite direction and partially overlaps (18 bp) SMU.747. SMU.747 and SMU.746 encode a putative permease (303 amino acids [aa]) and a substrate binding protein (271 aa), respectively (see Table S2 in the supplemental material), while SMU.745, called also lmrB, encodes an ATP-type multidrug resistance transporter (463 aa). The putative gene products appear to be membrane associated, with 7, 2, and 12 predicted transmembrane domains in SMU.747, SMU.746, and SMU.745, respectively. Unidirectional transcription and similar functions might suggest that these three genes may constitute an operon. However, the TransTermHP web service (http://transterm.cbcb.umd.edu/query.php) revealed the presence of a rho-independent terminator (Term 805), and some hairpin structures, which may cause transcript termination, were identified immediately downstream of the SMU.746 gene (data not shown). Analysis of the intergenic region between SMU.746 and SMU.745 (109 bp) with a promoter finding algorithm (BPROM; Softberry) revealed the presence of a putative promoter sequence in front of SMU.745 (data not shown). To confirm the presence of the promoter, a 163-bp DNA fragment including the intergenic region was cloned in front of a promoterless gusA gene in the pIB107 plasmid, resulting in the pIBL31 plasmid. A weak GusA activity (∼5 Miller units) suggests that SMU.745 can form a separate transcription unit. The low promoter activity of that construct might be due to the presence of a RAT (ribonucleic antitermination) structure similar to that identified in the bglP promoter (data not shown) (39). Further confirmation that SMU.746-SMU.747 forms a two-gene operon came from a genome context analysis of that locus (Fig. 2). Although the SMU.746-SMU.747 genes are conserved among different streptococci, the genomic context is quite different. None of the five genomes contained the SMU.745 gene in close vicinity (Fig. 2). Furthermore, in other oral streptococci, such as S. gordonii, S. mitis, and S. sanguinis, an SMU.745 homologue was not located near the SMU.746-SMU.747 locus (data not shown). Protein similarity searches revealed that only two systems, from S. ratti and S. macacae, had 80 to 90% identity, while most other systems were similar at more than 70% (see Table S2). However, because the SMU.746-SMU.747 genes are well conserved, they might have some common functions in streptococci.
FIG 2.
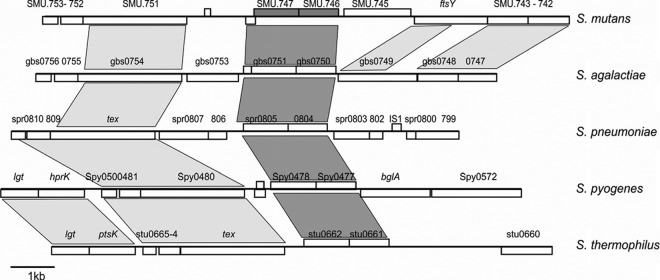
Synteny among the genomic regions of S. mutans (GenBank accession no. AE014133), Streptococcus agalactiae (AL735626), Streptococcus pneumoniae (AE007317), Streptococcus pyogenes (AE004092), and Streptococcus thermophilus (CP000023) containing SMU.745-SMU.747 homologs, indicated by dark gray. Syntenic regions are shown with gray boxes.
SMU.746-SMU.747 but not SMU.745 is involved in growth at low pH.
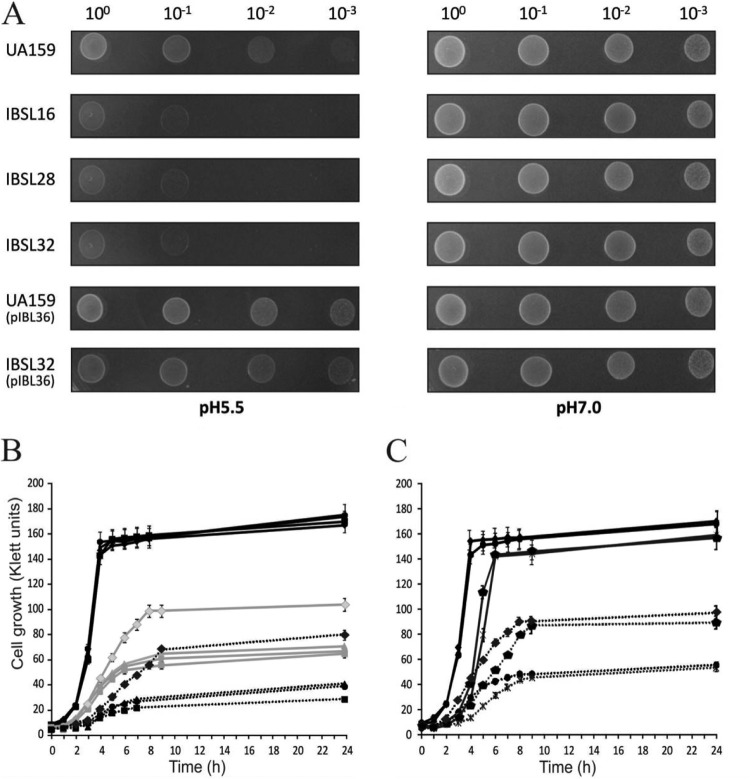
To confirm the role of these three genes in acid resistance, we constructed single-gene deletions for each of the three open reading frames (IBSL16, IBSL28, and IBSL23, respectively) and a double deletion mutant of SMU.746-SMU.747 (IBSL32) (Fig. 1; Table 1). Growth assays on THY pH 5.5 plates revealed that both SMU.746 and SMU.747 mutants were sensitive to low pH, while the sensitivity of the SMU.745 mutant was unchanged compared to that of the wild-type S. mutans (WT) strain (Fig. 3A and data not shown). Growth comparison of the WT and mutants in THY medium buffered to pH 7.0, 5.5, and 5.0 revealed that the single mutants and the SMU.746-SMU.747 double mutant were similar at pH 7.0 (Fig. 3A and B). However, at lower pH, the mutants grew much slower and reached the plateau at the lower cell densities of 62.1% ± 5.6% and 45.4% ± 8% of that of the WT strain at pHs 5.5 and 5.0, respectively (Fig. 3B). The ability of IBSL32 to grow at low pH was fully restored by complementation with the wild-type SMU.746-SMU.747 genes expressed from the P23 promoter in the plasmid pIBL36 (Fig. 3A and C). Although the growth rate and the final cell density were similar for the WT and the complemented mutants, we observed a slightly longer lag phase in strains carrying the complementing plasmid (Fig. 3C).
FIG 3.
(A) Effect of low pH on growth on THY plates. (B) Growth kinetics of the S. mutans mutants and the parental UA159 strains in liquid medium. (C) IBSL32 with the pIBL36 or pIB184Km plasmid in liquid medium. For growth on pH 5.0 and 7.0 THY agar plates, diluted overnight cultures (10 μl) were spot dried and plates were inoculated for 48 h at 37°C in 5% CO2. Growth kinetics was measured as described in Materials and Methods. Strains were grown in THY medium at pH 5.0, 5.5, and 7.0 (marked as black dotted, gray, and black lines, respectively). Samples are as follows: S. mutans UA159 (diamonds), IBSL16 (squares), IBSL28 (triangles), IBSL32 (circles), IBSL32/pIBL36 (pentagons), and IBSL32/pIB184Km (asterisks).
SMU.746-SMU.747 are not involved in superoxide or thermal stress responses.
In order to check if the SMU.746-SMU.747 genes have a pleiotropic effect on general stress responses in S. mutans, we conducted plate assays using common inductors of superoxide stress responses, such as methyl viologen and H2O2, as well as puromycin (which causes premature chain termination during protein synthesis), to mimic thermal stress. Our data showed no differences in growth of the SMU.746-SMU.747 mutants in the presence of those stressors in comparison to that of the WT strain (data not shown).
Inactivation of the SMU.746-SMU.747 genes affects biofilm formation in a medium- and pH-dependent manner.
Since biofilm formation is a well-known mechanism of survival and persistence in natural ecosystems, we were interested to see if SMU.746-SMU.747 mutation affected the ability to form biofilm. We observed that in THY medium with a neutral pH, all the mutant strains and the WT strain form similar amounts of biofilm (Fig. 4A and data not shown). However, at low pH, the amount of biofilm for the WT strain was slightly lower than that at the neutral pH. In contrast, to our surprise, all the mutants showed a 3.2- to 4.8-fold increase in biofilm formation (Fig. 4A and data not shown). The wild-type phenotype was restored in IBSL32 carrying the complementing pIBL36 plasmid.
FIG 4.
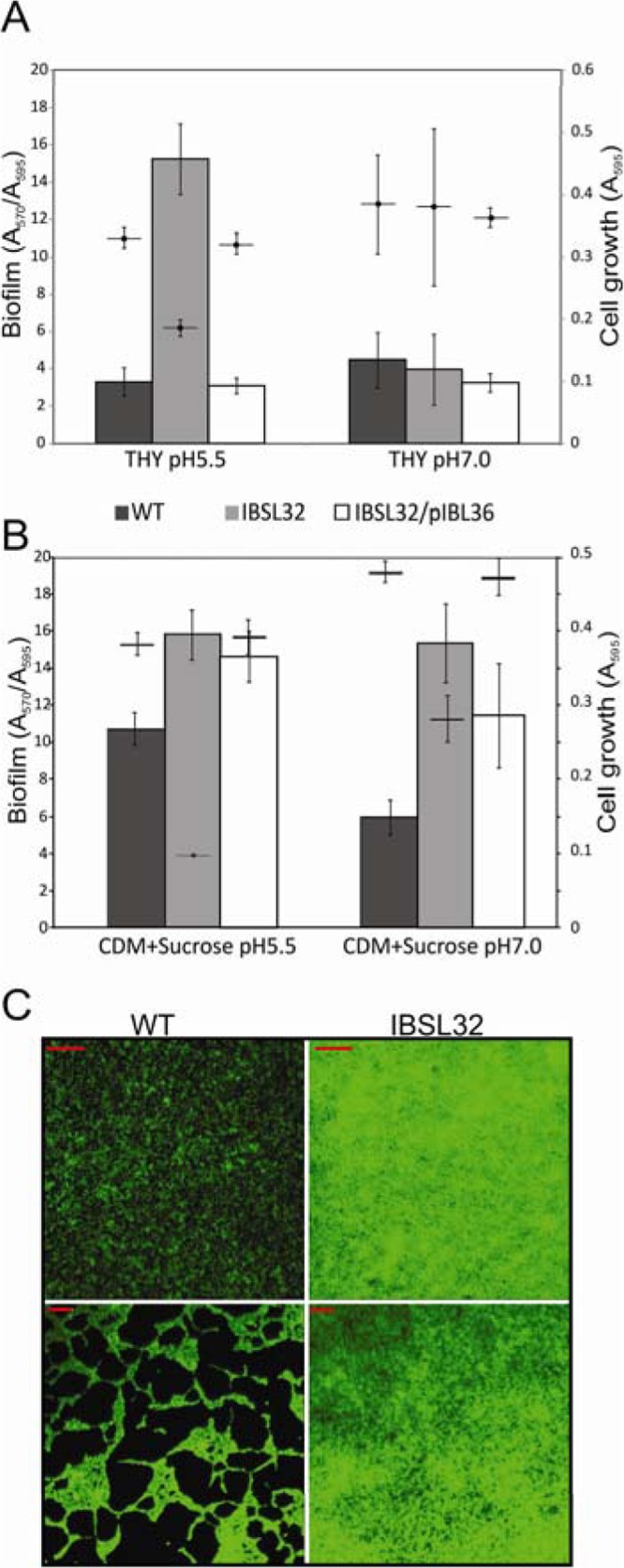
Biofilm formation of S. mutans UA159 and the SMU.746-SMU.747 mutant in THY (A) or CDM plus 1% sucrose (B) medium. Amounts of biofilm formation (A570/A595) are shown as black, gray, and white bars for the wild-type, IBSL32, and IBSL32/pIBL36 strains, respectively; and horizontal lines mark planktonic growth. The data are averages with standard deviations of the results from at least four independent experiments. (C) Microscopy photographs of 2-day-old biofilm formed by S. mutans UA159 (left panel) and the SMU.746-SMU.747 mutant strain (right panel) in THY pH 5.5 medium (magnification, ×10 [upper panel] or ×100 [lower panel]); red lines represent 100 and 10 μm for the upper and the lower panels, respectively.
Furthermore, in CDM medium with 1% sucrose, the IBSL32 mutant formed from 1.6- to 2.5-fold more biofilm than the WT strain in the corresponding medium (Fig. 4B). We noticed that the presence of the complementing plasmid pIBL36 not only restored the WT phenotype but also induced biofilm formation by 1.4- to 2.0-fold depending on pH (Fig. 4B).
Microscopic analysis of 2-day-old biofilm grown at pH 5.5 showed that the WT strain formed much less prolific biofilms, in which cells were scattered on the substratum as chains or aggregates that were generally very small and did not grow well (Fig. 4C, left panel, and data not shown). In contrast, the IBSL32 mutant strain covered more than 96% of the surface, forming an amorphic biofilm, although the average thickness was similar to that of the WT strain (Fig. 4C, right panel, and data not shown).
SMU.746-SMU.747 mutants are not defective in a major acid tolerance mechanism.
A major acid resistance mechanism involves F1-Fo ATPase activity. Although no typical ABC transporter motif (ATP binding sites) has been found, an extensive BLAST search for the SMU.747 protein revealed that the region at 129-GAIIVAISLGILLGFIVDE-147 showed some similarities to the F1-Fo ATPase domain. Since three glycine residues responsible for ATP binding were highly conserved, we assumed that IBSL28 or IBSL32 mutants might be defective in F1-Fo ATPase activity. However, the ATPase activity showed no statistically significant difference between the WT strain and IBSL32 mutant (data not shown). The mutant strain showed a slightly higher activity that might be due to the induction of F1-Fo ATPase activity when the SMU.746-SMU.747 genes are absent.
The SMU.746-SMU.747 mutation does not affect membrane proton permeability.
The bacterial membrane plays an important role in the aciduricity of S. mutans. A previous study demonstrated that S. mutans possesses a membrane that is rather impermeable to protons (40). A change in the membrane composition leads to bacteria sensitive to low pH. Since both SMU.746 and SMU.747 are integral membrane proteins, the deletion of either or both could change membrane stability and make it more permeable to protons. If the mutant strains had altered proton permeability and could not maintain a normal ΔpH across the membrane, that could account for the observed acid sensitivity phenotype. Therefore, we measured the proton permeability of the WT and the IBSL32 mutant strains. The rise in external pH in both cases was almost identical, with the final pH after cell permeabilization slightly higher in the mutant strain (data not shown). These results suggest that membrane permeability may not be responsible for the impaired growth of the IBSL32 mutant at a lower pH.
SMU.746-SMU.747 mutants survive both short-term acid shock and a long-term acid survival.
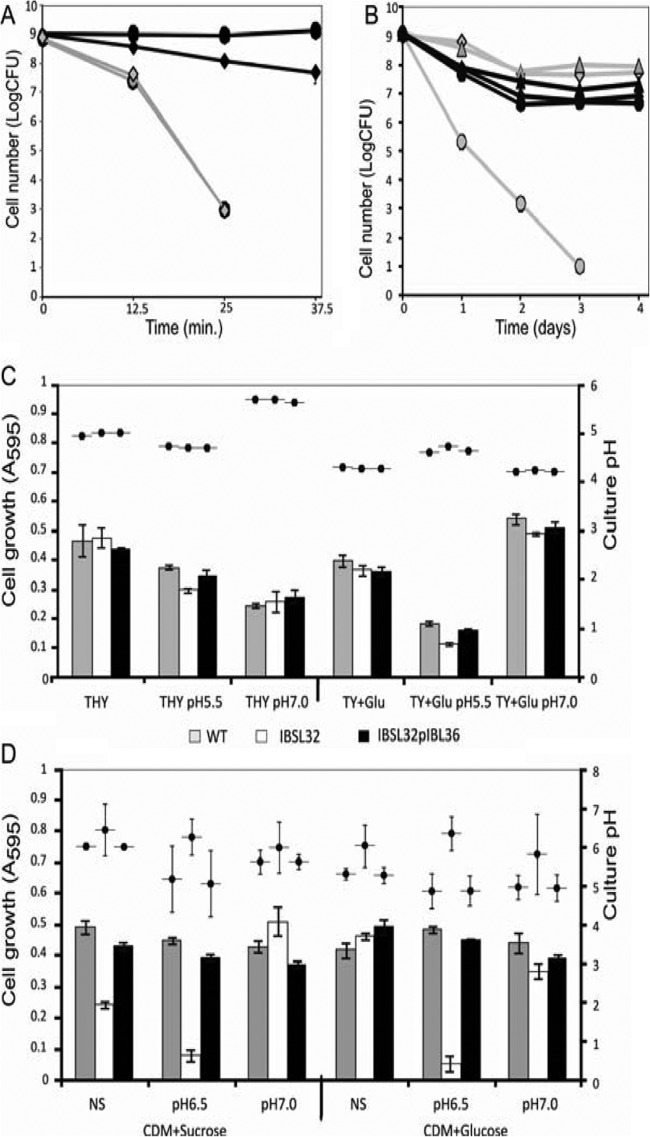
A higher difference in growth ability between the IBSL32 and WT strains at pH 5.0, as shown in Fig. 3, suggested that lower pH reduces SMU.746-SMU.747 mutant viability. To check the effect of low pH on survival, both the WT and double mutant strains were incubated in a low-pH buffer (pH 2.8) for up to 50 min, and cell viability was measured. Results showed that the two strains had the same killing rate over time, and the 25 min of incubation was sufficient for a million-fold reduction in cell viability (Fig. 5A).
FIG 5.
Effect of acid stress on cell survival and acidogenesis of S. mutans strains in different media. (A) Acid killing of S. mutants UA159 (diamonds) and IBSL32 mutant (circles) strains. Aliquots were plated on THY agar upon suspension in 0.1 M glycine (pH 2.8, gray lines; pH 7.0, black lines) and each 12.5 min thereafter. (B) A long-term survival assay was carried out as described in Materials and Methods. Aliquots of overnight cultures were plated daily on THY agar. The data are the averages with standard deviations of the results from two independent experiments. Black lines represent THY plus 1% glucose; gray lines are CDM plus 1%glucose. WT, IBSL32, and IBSL32pIBL36 strains are marked as diamonds, circles, and triangles, respectively. (C) Growth and acid production in THY and TY plus 1% glucose measured in 18-h cultures. (D) Growth and acid production in CDM with 1% glucose or sucrose in 2-day-old cultures. The data represent average values of 5 independent experiments.
To assess the ability of S. mutans strains to survive under long-term acid stress, we used two types of media: TY medium with 1% glucose, as described earlier (36), and CDM with 1% glucose as a sole carbon source. In the presence of glucose, S. mutans uses glycolysis to produce lactic acid and is able to reduce the pH from 7.0 to 3.7 within 30 min (41). Therefore, culturing bacteria in this medium allows the long-term effects of low pH on cell survival to be studied. The data showed that in TY medium, the wild-type strain had almost the same death rate as the IBSL32 mutant. However, in CDM, viability of the IBSL32 mutant strain was 3 to 6 orders of magnitude lower than that of the WT strain in the following days (Fig. 5B).
SMU.746-SMU.747 mutants do not acidify minimal medium in the presence of sugars.
The growth of the IBSL32 mutant in CDM was strongly affected by low pH, and it was not able to grow at <pH 6.5, especially with simple sugars as a carbon source (data not shown). Prolonged incubation of all strains in CDM plus glucose medium and CDM plus glucose with an initially stabilized pH at 6.5 or 7.0 showed that the SMU.746-SMU.747 mutant was able to grow only when the initial pH was set above 7.0. If the initial pH of the CDM was 6.5, the growth of IBSL32 was limited (Fig. 5D).
Although acidogenesis in sugar-containing medium was expected to occur, we checked if our strains were indeed able to reduce the external pH. In rich THY medium, after 18 h of incubation, all strains acidified medium to the same final pH. Depending on the initial pH, the values ranged from 4.97 ± 0.03, 4.72 ± 0.01, and 5.62 ± 0.03 in not-stabilized, pH 5.5, and pH 7.0 media, respectively (Fig. 5C). In the case of TY plus glucose medium, slightly lower but still similar pH values were observed in the case of not-stabilized and neutral initial pHs (4.28 ± 0.02, and 4.21 ± 0.01). The lower value at neutral initial pH correlated with a higher cell density for all strains (A595 of 0.51 ± 0.01) in comparison to that with the not-stabilized medium (A595 of 0.376 ± 0.01). In medium stabilized at ph5.5, growth was significantly reduced for all strains compared to that at pH 7.0. However, growth of the IBSL32 mutant strain was slightly lower than that of the WT and complemented strains (A595 of 0.12 ± 0.005 versus 0.18 ± 0.007 or 0.16 ± 0.005) (Fig. 5C). These growth differences were responsible for the observed pH values; pH 4.62 ± 0.02 for the WT and IBSL32/pIBL36 strains and pH 4.78 ± 0.04 for the mutant strain (Fig. 5C).
In CDM with 1% sucrose or glucose, the WT and complemented IBSL32 mutant strains lowered the external pH in a medium-dependent fashion, while the mutant strain was not able to acidify culture media (Fig. 5D). The strongest effect observed in CDM plus glucose medium with an initial pH of 6.5 correlated with the minimal growth of mutant strains in that medium (Fig. 5D). But the results obtained in CDM plus glucose and sucrose, unbuffered, and pH 7.0 buffered media, where differences in cell growth after 42 h were negligible, were similar (Fig. 5D). Since the major product that causes the pH reduction in media with high sugar concentrations is lactic acid, we speculated that an absence of SMU.746-SMU.747 affected the amount of lactic acid produced.
The SMU.746-SMU.747 mutant has lower glycolytic activity.
Lack of the reduction of external pH in glucose-containing medium could be due to either a deficiency in lactic acid production or an ability to export the lactate outside the cell. To assess the ability of our strains to produce lactic acid, we ran a standard glycolysis pH drop-down experiment. The data showed that in pH 7.0 buffer, all three strains were able to reduce pH to ∼4.1 during 2 h of incubation, although the kinetics of the pH drop was slightly lower in the case of the IBSL32 mutant (Fig. 6A). The situation was different when cells were resuspended in pH 5.0 buffer. The complemented IBSL32/pIBL36 strain showed the fastest pH drop; however, the final pH was almost identical to that of the WT strain. On the other hand, the kinetics of the pH drop was lower in the IBSL32 mutant, and the final pH was 0.25 units higher than that of the WT strain (Fig. 6B). These data suggest that an absence of SMU.746-SMU.747 leads to lower glycolytic activity.
FIG 6.
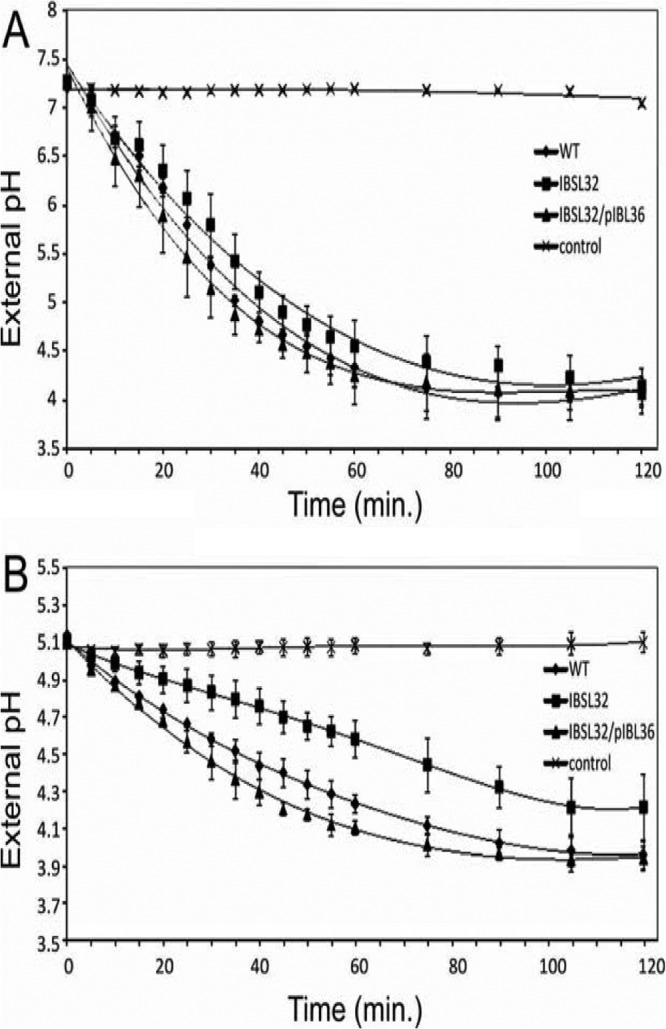
Drop of external pH in S. mutans cell suspensions after induction with 1% glucose. Cells were resuspended in 0.05 M phosphate-citrate buffer, pH 7.0 (A) or pH 5.0 (B). Samples are as follows: S. mutans UA159 (WT), IBSL32, and complemented IBSL32. Controls represent average pH values for all 3 strains without induction. The data are averages with standard deviations of results from three independent experiments.
SMU.746-SMU.747 permease is responsible for amino acid transport.
To identify the function of the SMU.746-SMU.747 permease, we attempted to substitute for some of the CDM components. A cell can take up amino acids as individual residues or as small peptides, both carried out by different transport mechanisms. When amino acids in CDM were replaced by peptone (20 mg/ml), we observed no difference in growth at pH 7.0 between the WT and the mutant strain after 24 h (A595 of 0.99 ± 0.03 and 0.98 ± 0.06, respectively). In contrast, at pH 6.5, the WT strain grew much better than the IBSL32 mutant strain (A595 of 1.05 ± 0.03 versus 0.11 ± 0.14, respectively). At the same pH, growth of the IBSL32 mutant in CDM supplemented with amino acids was minimal (A595 of 0.046 ± 0.0026). These data indicate that that the SMU.746-SMU.747 system might be responsible for uptake of some amino acids. We also noticed that the growth capacity of the mutant strain in CDM at pH 6.5 depended on the conditions of cells used for inoculation. Growth of starved cells from a late-stationary-phase culture was severely reduced or stopped altogether, while highly energized cells from an exponential-phase culture grew much better, suggesting that the role of the SMU.746-SMU.747 permease is increased under energy-deprived conditions.
DISCUSSION
S. mutans employs multiple mechanisms to mount acid tolerance responses when the organism is exposed to a low-pH environment. Among them, the most important ones are increased expression of proton pumps (40), protection and repair of macromolecules, changes in cell membrane permeability (13), production of alkali by the agmatine deiminase system (17), and alteration of metabolic pathways (42). Consistent with the notion, Gong and colleagues (43) have shown that low pH can induce about 14% of the S. mutans genes when the bacterium is exposed to pH 5.5. In addition to genes that encode proteins involved in the above-mentioned cellular functions needed for the acid tolerance response, about one-fifth of the genes encode hypothetical proteins whose functions are unknown. With the help of a powerful transposon mutagenesis system, we identified two new genes, SMU.746 and SMU.747, that were not previously reported to be involved in aciduricity.
Both the SMU.746 and SMU.747 genes appeared to encode a membrane-associated permease complex. Though there is very little sequence similarity between the SMU.746-SMU.747 and F1-Fo ATPase proteins, a conserved motif is present in both the complexes. However, our data strongly suggest that the SMU.746-SMU.747 complex does not act as a proton pump. Growth studies with CDM supplemented with either peptone or individual amino acids suggest that SMU.746-SMU.747 may have a role in amino acid transport under low pH.
S. mutans has simple nutritional requirements and is able to grow with the components of saliva as its sole sources of carbon and nitrogen (44–46). Most of the strains can grow in minimal medium supplemented with only with cysteine and glutamine, and all amino acid biosynthetic pathways have been identified in the genome (22). Despite the fact that glutamate can be synthesized in vivo, five predicted ABC transporters involved in transport of that amino acid have been identified (22). A glutamate transporter encoded by the glnQHMP operon has been described as responsible for uptake of 95% glutamate (24). Surprisingly, deletion of the glnQHMP operon reduced the acid tolerance response in S. mutans (24). Furthermore, other amino acid biosynthesis and/or catabolism genes also play an important role in the acid tolerance response in S. mutans. For example, Santiago et al. (18) have recently shown that IlvE, a branched-chain amino acid aminotransferase required for catabolism of isoleucine and valine, is required for acid tolerance. IlvE either directly or indirectly regulates proton pumps, since an ilvE mutant strain exhibited a defect in F1-Fo ATPase activity. Thus, amino acids play a crucial role in the acid tolerance response in S. mutans.
It has long been observed that low pH reduces the growth of streptococci. We observed that the growth rate and the final cell density depended on the initial pH of the THY medium (Fig. 3). In this medium, mutation in a single transport system, especially for amino acid transport, should not affect the growth rate, since the nutrients can be taken up as peptides of various chain lengths by the corresponding oligopeptide transport system(s). At low pH, the activity of ATP-dependent transport systems is reduced to protect the organism from wasting metabolic energy (47). Specifically, under these conditions, the role of a specific transporter would be apparent and a mutation in the transporter-encoding gene would adversely affect the growth. Indeed, we observed that in THY and TY media, the differences in growth between the WT and mutant strains were negligible at neutral pH. The lack of growth differences indicated that SMU.746-SMU.747 mutants can adapt to low pH in the same way as the UA159 strain (37). Thus, low pH has no direct impact on mutant strain survival, as is evident from the long exposure and short-term acid killing experiments. In contrast, when the pH of the rich medium was ≤5.5, growth of the mutants was much more affected than that of the wild-type strain, confirming a specific role of SMU.746-SMU.747 permease under these conditions.
Although, as mentioned above, S. mutans can grow in minimal medium supplemented with cysteine and glutamate (44–46), the high metabolic activity and energy consumption should make a permease-deficient strain difficult to grow. In fact, we observed that in CDM the growth of the SMU.746-SMU.747 mutant was significantly affected by the initial pH. The mutant strain grew but much slower (40 to 48 h) and only when the initial pH was neutral (Fig. 5D). At lower pH, the cellular metabolism and energy level are lower due to repression of amino acid synthesis genes, but the energy consumption is higher due to pH gradient maintenance. Consistent with this concept, we observed that if the initial pH was ≤6.5, the mutant was not able to grow (Fig. 5D).
In S. mutans, the presence of simple sugars like glucose and fructose induces glycolysis and lactic acid production (6, 7). We noticed that in CDM, the IBSL32 mutant showed a reduced ability to lower the external pH compared to the wild type, UA159 (Fig. 5D). However, we observed a slight difference in glycolytic activity between the wild type and the IBSL32 mutant strain in buffered medium at pH 7.0 (Fig. 6A). The difference in the external pH drop between the wild type and the mutant strain was slightly greater in buffer at pH 5.0, although both strains were able to acidify the medium (Fig. 6B). Reduced glycolysis and lactic acid production were also observed in the case of the glutamate transporter mutant, as mentioned earlier (24). Since glutamate is the major precursor for other amino acid synthesis pathways, the relationship between amino acid transport and glycolysis is expected to be the same. Based on our current knowledge about the S. mutans metabolic pathways, we can speculate on why the SMU.746-SMU.747 deficiency affects lactic acid production. As previously reported (48), the amount of pyruvate, which is converted to lactate, is reduced by the amount necessary for amino acids production. An alternate explanation may involve NAD+-NADH. Both glycolysis and amino acid biosynthesis require NAD+ as a cofactor (49, 50). Since the total intracellular NAD+ pool is limited, competition between these two processes should slow down both pathways. Further studies are necessary to reveal the mechanism of interaction between amino acid transport and glycolysis.
The most surprising observation was that the SMU.746-SMU.747 mutant showed increased biofilm formation at low pH when planktonic growth was reduced (Fig. 4A). In rich medium, such as in THY, biofilm formation depended clearly on the initial pH of the medium, while in minimal medium we observed that the mutant strain could form biofilm more efficiently even at neutral pH (Fig. 4B). We also noticed that overall biofilm formation was better for all strains at a lower initial pH induced in minimal medium. At present, we can only speculate on why the SMU.746-SMU.747-deficient strain forms better biofilm than the wild type at low pH. As discussed earlier, the growth rate of the SMU.746-SMU.747 mutant in THY medium at pH 5.5 is much lower, suggesting that the cells suffered a lack of necessary nutrients. This nutritional stress is responsible for the induction of biofilm formation in the IBSL32 mutant. On the other hand, since in minimal medium the nutrients are limited, this automatically triggers biofilm formation even at neutral pH, and further acid stress increases enhanced biofilm formation. This hypothesis is in complete agreement with our observations that biofilm formation by the wild-type strain was increased ∼2-fold at lower pH compared with that at neutral pH (Fig. 4B).
In the natural environment, the energy supply for growth and survival is often a limiting factor, and many organisms are forced to scavenge energy from all potential sources. Organisms that regularly encounter such energy-limited conditions, such as oral streptococci, have developed solute transporters that can play crucial roles in these energy-scavenging processes. Our data are in agreement with this conclusion, since the SMU.746-SMU.747 mutant is impaired in growth under some stress conditions (low pH and minimal medium), when the cell energy stock is relatively low due to expenses for the maintenance of pH gradient and anabolic processes. Additional genetic repression of available transporters under these conditions increases the role of specific permeases, making them more important for cell survival in the natural environment. Further studies of membrane permeases in S. mutans are necessary to confirm their role in the physiology of that important pathogen.
Not only do lactic acid bacteria, including streptococci, acidify their environment, but most of the species are also exposed to a naturally acidic environment. For example, group B streptococci (GBS) often encounter acidic pH in the vaginal environment, where they often colonize, and a recent report suggests that clinical GBS isolates from humans form better biofilm and withstand acidic environment better than the nonclinical isolates (51). Thus, it is not surprising to find that most streptococci also encode the homologs of SMU.746-SMU.747. Although nothing is known about the function of these homologs, we speculate that these proteins (permeases) also play a role in acid tolerance response and transport processes.
Supplementary Material
ACKNOWLEDGMENT
This research was supported in part by an NIH-NIDCR grant (DE021664) to I.B.
Footnotes
Published ahead of print 18 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00960-13.
REFERENCES
- 1.Kestenbaum RC. 1968. Bacterial specificity in the etiology of caries and periodontal disease. J. Dent. Res. 47:925. 10.1177/00220345680470065501 [DOI] [PubMed] [Google Scholar]
- 2.Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh PD. 2005. Dental plaque: biological significance of a biofilm and community life-style. J. Clin. Periodont. 32(Suppl 6):7–15. 10.1111/j.1600-051X.2005.00790.x [DOI] [PubMed] [Google Scholar]
- 4.Banas JA, Vickerman MM. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89–99. 10.1177/154411130301400203 [DOI] [PubMed] [Google Scholar]
- 5.Lee SF, Progulske-Fox A, Erdos GW, Piacentini DA, Ayakawa GY, Crowley PJ, Bleiweis AS. 1989. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II). Infect. Immun. 57:3306–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimura J, Saito T, Yoneyama H, Lan Bai L, Okumura K, Isogai E. 2012. Biofilm formation by Streptococcus mutans and related bacteria. Adv. Microbiol. 2:208–215. 10.4236/aim.2012.23025 [DOI] [Google Scholar]
- 7.Abbe K, Takahashi S, Yamada T. 1982. Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J. Bacteriol. 152:175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quivey RG, Kuhnert WL, Hahn K. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301–314. 10.1177/10454411010120040201 [DOI] [PubMed] [Google Scholar]
- 9.Matsui R, Cvitkovitch D. 2010. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 5:403–417. 10.2217/fmb.09.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter PD, Hill C. 2003. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429–453. 10.1128/MMBR.67.3.429-453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton IR, Svensater G. 1998. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol. Immunol. 13:292–300. 10.1111/j.1399-302X.1998.tb00710.x [DOI] [PubMed] [Google Scholar]
- 12.Len AC, Harty DW, Jacques NA. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339–1351. 10.1099/mic.0.27008-0 [DOI] [PubMed] [Google Scholar]
- 13.Fozo EM, Quivey RG., Jr 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70:929–936. 10.1128/AEM.70.2.929-936.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna MN, Ferguson RJ, Li YH, Cvitkovitch DG. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183:5964–5973. 10.1128/JB.183.20.5964-5973.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn K, Faustoferri RC, Quivey RG., Jr 1999. Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol. Microbiol. 31:1489–1498. 10.1046/j.1365-2958.1999.01292.x [DOI] [PubMed] [Google Scholar]
- 16.Sheng J, Baldeck JD, Nguyen PTM, Quivey RG, Marquis RE. 2010. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can. J. Microbiol. 56:539–547. 10.1139/W10-039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griswold AR, Jameson-Lee M, Burne RA. 2006. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188:834–841. 10.1128/JB.188.3.834-841.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santiago B, MacGilvray M, Faustoferri RC, Quivey RG., Jr 2012. The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J. Bacteriol. 194:2010–2019. 10.1128/JB.06737-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welin-Neilands J, Svensater G. 2007. Acid tolerance of biofilm cells of Streptococcus mutans. Appl. Environ. Microbiol. 73:5633–5638. 10.1128/AEM.01049-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745. 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- 21.Mah T-FC, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39. 10.1016/S0966-842X(00)01913-2 [DOI] [PubMed] [Google Scholar]
- 22.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas S, Biswas I. 2011. Role of VltAB, an ABC transporter complex, in viologen tolerance in Streptococcus mutans. Antimicrob. Agents Chemother. 55:1460–1469. 10.1128/AAC.01094-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krastel K, Senadheera DB, Mair R, Downey JS, Goodman SD, Cvitkovitch DG. 2010. Characterization of a glutamate transporter operon, glnQHMP, in Streptococcus mutans and its role in acid tolerance. J. Bacteriol. 192:984–993. 10.1128/JB.01169-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korithoski B, Krastel K, Cvitkovitch DG. 2005. Transport and metabolism of citrate by Streptococcus mutans. J. Bacteriol. 187:4451–4456. 10.1128/JB.187.13.4451-4456.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luz DE, Nepomuceno RS, Spira B, Ferreira RC. 2012. The Pst system of Streptococcus mutans is important for phosphate transport and adhesion to abiotic surfaces. Mol. Oral Microbiol. 27:172–181. 10.1111/j.2041-1014.2012.00641.x [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Senadheera DB, Lévesque CM, Cvitkovitch DG. 2012. TcyR regulates L-cystine uptake via the TcyABC transporter in Streptococcus mutans. FEMS Microbiol. Lett. 328:114–121. 10.1111/j.1574-6968.2011.02492.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas I, Drake L, Biswas S. 2007. Regulation of gbpC expression in Streptococcus mutans. J. Bacteriol. 189:6521–6531. 10.1128/JB.00825-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Biswas I. 2009. 3′-Phosphoadenosine-5′-phosphate phosphatase activity is required for superoxide stress tolerance in Streptococcus mutans. J. Bacteriol. 191:4330–4340. 10.1128/JB.00184-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. 2005. WebACT—an online companion for the Artemis Comparison Tool. Bioinformatics 21:3665–3666. 10.1093/bioinformatics/bti601 [DOI] [PubMed] [Google Scholar]
- 31.Hossain MS, Biswas I. 2012. An extracelluar protease, SepM, generates functional CSP in Streptococcus mutans UA159. J. Bacteriol. 194:5886–5896. 10.1128/JB.01381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. 10.1099/mic.0.2008/019265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biswas S, Biswas I. 2006. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans. J. Bacteriol. 188:988–998. 10.1128/JB.188.3.988-998.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao L, Biswas I. 2013. ClpL is required for folding of CtsR in Streptococcus mutans. J. Bacteriol. 195:576–584. 10.1128/JB.01743-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen ZT, Burne RA. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682–2691. 10.1128/JB.186.9.2682-2691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 192:2546–2556. 10.1128/JB.00028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belli WA, Marquis RE. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd DA, Cvitkovitch DG, Bleiweis AS, Kiriukhin MY, Debabov DV, Neuhaus FC, Hamilton IR. 2000. Defects in D-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182:6055–6065. 10.1128/JB.182.21.6055-6065.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cote CK, Honeyman AL. 2006. Transcriptional analysis of the bglP gene from Streptococcus mutans. BMC Microbiol. 6:37. 10.1186/1471-2180-6-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bender GR, Sutton SV, Marquis RE. 1986. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect. Immun. 53:331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fozo EM, Quivey RG., Jr 2004. The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J. Bacteriol. 186:4152–4158. 10.1128/JB.186.13.4152-4158.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247–3255. 10.1099/mic.0.2008/023770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong Y, Tian XL, Sutherland T, Sisson G, Mai J, Ling J, Li YH. 2009. Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation. Microbiology 155:3322–3332. 10.1099/mic.0.031591-0 [DOI] [PubMed] [Google Scholar]
- 44.Terleckyj B, Shockman GD. 1975. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlsson J. 1970. Nutritional requirements of Streptococcus mutans. Caries Res. 4:305–320. 10.1159/000259653 [DOI] [PubMed] [Google Scholar]
- 46.St Martin EJ, Wittenberger CL. 1980. Regulation and function of ammonia-assimilating enzymes in Streptococcus mutans. Infect. Immun. 28:220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poolman B, Driessen AJ, Konings WN. 1987. Regulation of solute transport in streptococci by external and internal pH values. Microbiol. Rev. 51:498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Len AC, Harty DW, Jacques NA. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology 150:1353–1366. 10.1099/mic.0.26888-0 [DOI] [PubMed] [Google Scholar]
- 49.Garvie EI. 1980. Bacterial lactate dehydrogenases. Microbiol. Rev. 44:106–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cvitkovitch DG, Gutierrez JA, Bleiweis AS. 1997. Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans. J. Bacteriol. 179:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho YR, Li CM, Yu CH, Lin YJ, Wu CM, Harn IC, Tang MJ, Chen YT, Shen FC, Lu CY, Tsai TC, Wu JJ. 2013. The enhancement of biofilm formation in group B streptococcal isolates at vaginal pH. Med. Microbiol. Immunol. 202:105–115. 10.1007/s00430-012-0255-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.