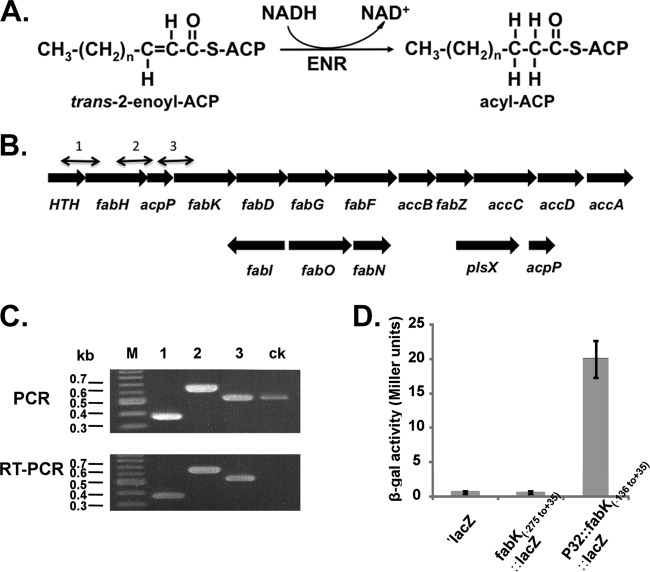
FIG 1.
The ENR reaction, the fabK and fabI genome neighborhoods, and transcriptional analyses of the operon containing fabK. (A) The enoyl-acyl-ACP reductase reaction. (B) Genetic organization of the E. faecalis fatty acid biosynthetic genes (fab). The numbered short lines (1, 2, and 3) represent the specific PCR amplicons observed in the PCR and RT-PCR assays shown in panel C. The second acpP gene (EF_3111 in strain V583) located downstream of plsX is conserved in all extant E. faecalis genome sequences. (C) PCR and RT-PCR analyses of the fab operon. The results for the four genes at the 5′ end of the operon are shown. The helix-turn-helix-encoding gene probably encodes a homologue of the FabT transcription factor found in other Firmicutes bacteria (43, 44). The primer numbering system is the same as that for panel B. Both PCR products obtained from genomic DNA and those obtained by RT-PCR were separated by electrophoresis on a 1.5% agarose gel. The ck designation denotes two neighboring genes transcribed from the opposite strand that were included as controls. (D) β-Galactosidase activities of fabK::lacZ translational fusions. The wild-type strain FA2-2 carrying each of the fusion plasmids was grown in GM17 medium to mid-log phase, and β-galactosidase activities were measured in more than three independent experiments. The error bars indicate standard deviations. The plasmids carried a promoterless lacZ (′lacZ), the −275 to +35 fragment, which includes the first 35 bp of the fabK coding sequence fused to lacZ (fabK-lacZ), or the −136 to +35 fragment driven by the P32 promoter. The fusion plasmids were pBHK322, pBHK394, and pBHK323, respectively.