FIG 4.
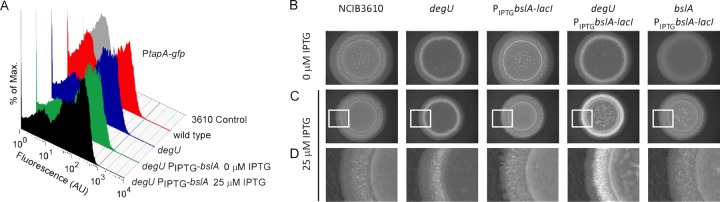
The degU mutant biofilm architecture does not influence matrix gene expression. (A) Flow cytometry analysis of cells grown under biofilm formation conditions for 17 h for strains NRS2394 (3610 sacA-PtapA-gfp), NRS2745 (degU sacA-PtapA-gfp), and NRS3067 (degU sacA-PtapA-gfp amyE-PIPTG-bslA-lacI) in the presence or absence of IPTG, as indicated. Strain 3610 was used as a nonfluorescent control. Fluorescence on the x axis is in arbitrary fluorescent units (AU). The data sets are representative of the results observed in three independent experiments. (B to D) Biofilm morphology, as indicated by the formation of a complex colony on MSgg agar after 17 h, for wild-type strain NCIB3610 (3610), a degU strain (NRS1314), NCIB3610 carrying amyE-PIPTG-bslA-lacI (NRS2297), degU amyE-PIPTG-bslA-lacI) (NRS2298), and bslA amyE-PIPTG-bslA-lacI (NRS2299). (B) 0 μM IPTG; (C) 25 μM IPTG; (D) higher-magnification images of the regions highlighted in panel C.
