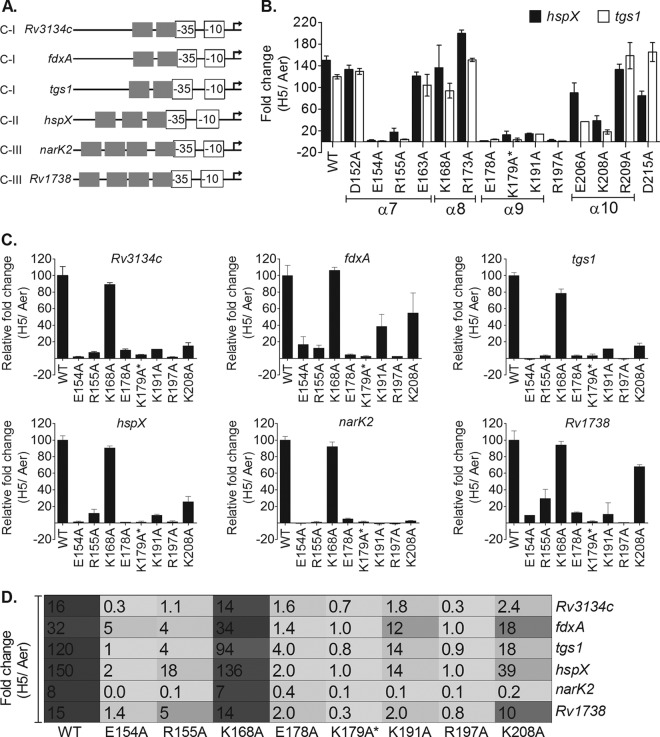
FIG 2.
Identification of transcription activation-defective devR mutants. (A) Conserved promoter architecture of regulon promoters. The overlap of the −35 element with the TSP-proximal Dev box is shown (15–17, 27). Experimentally determined hypoxic transcription start points (TSPs) are indicated with bent arrows. C-I, C-II, and C-III refer to class I, class II, and class III promoters, respectively. (B) M. tuberculosis mutant strains were screened for tgs1 and hspX transcripts by RT-qPCR analysis. The fold change in transcripts under 5-day “hypoxic” versus “aerobic” (H5/Aer) conditions is shown. (C) Selected mutant strains (from panel B) were further tested for their activation properties on class I (Rv3134c, fdxA, and tgs1), class II (hspX), and class III (narK2 and Rv1738) promoters by RT-qPCR. Relative fold change values (H5/Aer) in mutants with respect to WT DevR (taken as 100) are shown. (D) Comparison of fold activation (H5/Aer) of various regulon promoters in WT and mutant M. tuberculosis strains as measured by RT-qPCR analysis.