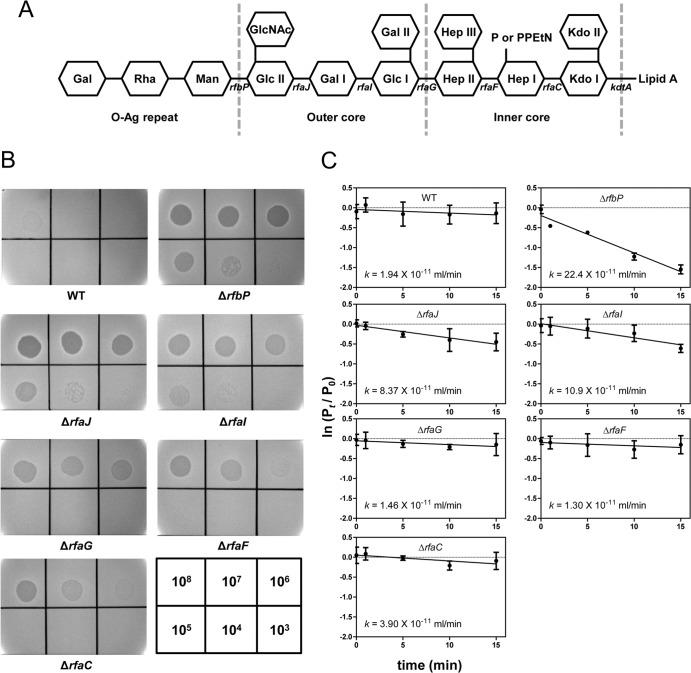
FIG 3.
SSU5 adsorbed to the outer core OS of Salmonella LPS. (A) Schematic representation of the LPS structure of S. Typhimurium. Carbohydrates are shown as hexagons, and the relevant genes involved in the biosynthesis of various residues of LPS are indicated below the line connecting the hexagon. The mutation of each gene caused the elimination of the left residues marked with gene names on the structure (rfbP, undecaprenyl-phosphate galactosephosphotransferase; rfaJ, LPS 1,2-glycosyltransferase; rfaI, LPS 1,3-galactosyltransferase; rfaG, glucosyltransferase I; rfaF, ADP-heptose-LPS heptosyltransferase; rfaC, LPS heptosyltransferase I; kdtA, 3-deoxy-d-manno-octulosonic acid transferase). Abbreviations: Kdo, 3-deoxy-d-manno-octulosonic acid; Hep, heptose; Glc, glucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Man, mannose; Rha, rhamnose; P, phosphate; PPEtN, pyrophosphorylethanolamine. (B) SSU5 spotting assay with mutants with truncations in the LPS core OS. Serially diluted (10-fold) SSU5 lysates were spotted on the lawns of indicated Salmonella mutants. The phage titer (PFU/ml) used for each spot is indicated in the grids at the bottom right corner. (C) Assay of adsorption of SSU5 to various core OS mutants. The adsorption constant (k) was calculated as described in the text. The means with SD of three independent assays are represented.