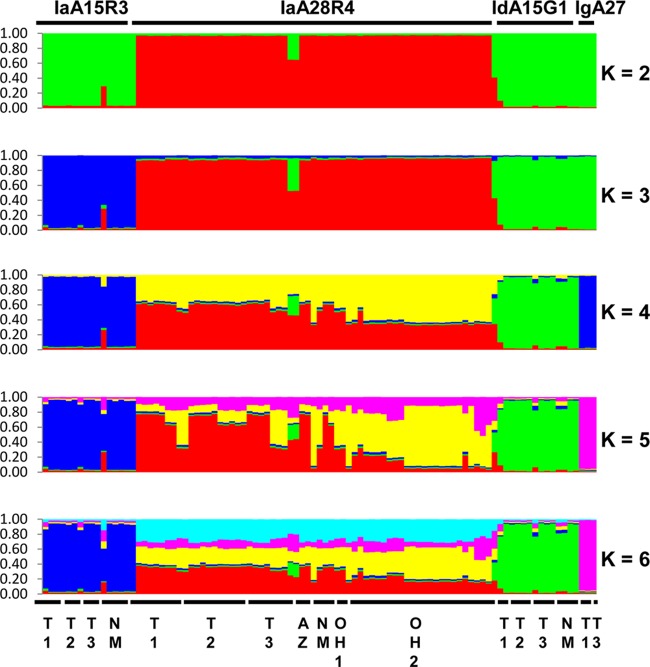
FIG 1.
Population substructure of 95 specimens of C. hominis from Ohio, Texas, New Mexico, and Arizona in the United States by Bayesian analyses of the MLST allelic data. Subpopulation patterns are shown for K values of 2 to 6 used in the analysis. Colored regions indicate major ancestral contributions. Mixed subtypes are indicated by the pattern of color combinations. Subtype identities of the specimens at the gp60 locus are shown above the color patterns, and the geographic origins of the specimens are shown below the color patterns. T1 and T2, specimens from an outbreak in two neighboring northeastern counties in Texas; T3, specimens from sporadic cases in a central Texas city; NM, specimens from sporadic cases (except for two IaA15R3 specimens) from New Mexico; AZ, specimens from an outbreak in Arizona; OH1 and OH2, specimens from two outbreaks in central Ohio. All specimens are from 2008, except for OH1, which represents two specimens from 2005.