FIG 3.
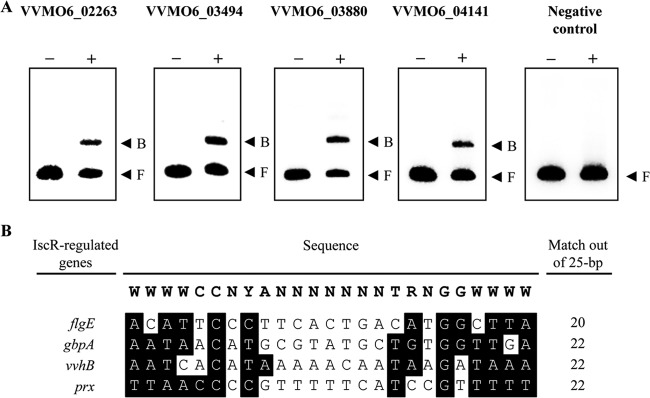
Verification of IscR binding to the regulatory region of the newly identified IscR regulon. (A) The regulatory regions of VVMO6_02263 (flgE), VVMO6_03494 (gbpA), VVMO6_03880 (vvhB), or VVMO6_04141 (prx) and a part of the iscR coding region (negative control) were radioactively labeled and then used as probe DNAs. The same locus tag numbers that appear in Fig. 2 are at the top of each panel. −, without IscR; +, with IscR; B, bound DNA; F, free DNA. (B) Comparison of the consensus E. coli IscR-binding sequence and the potential V. vulnificus IscR-binding sequences. The consensus E. coli IscR-binding sequence (type 2) (38) is shown at the top. Sequences from the regulatory regions of flgE, gbpA, vvhB, and prx, retrieved from GenBank (accession numbers CP002469 and CP002470), are aligned below. Individual bases identical to those conserved in the consensus E. coli IscR-binding sequence are highlighted. Numbers of bases that match those of the consensus E. coli IscR-binding sequence are indicated on the right. R, A or G; Y, C or T; W, A or T; N, any base.
