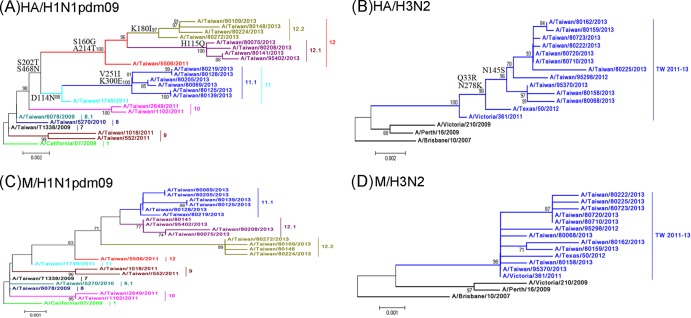
FIG 1.
Phylogenetic relationships of the full-length HA gene (A and B) and the M gene (C and D) sequences of the influenza A(H1N1)pdm09 and A(H3N2) viruses in Taiwan. The phylogenetic trees were constructed using the neighbor-joining method, with 1,000 bootstrap replications. Branch values of >70 are indicated. The sequences of the vaccine strains, including that of A/California/7/2009 of the A(H1N1)pdm09 subtype, as well as A/Texas/50/2012, A/Victoria/361/2011, A/Victoria/210/2009, A/Perth/16/2009, and A/Brisbane/10/2007 of the A(H3N2) subtype, were downloaded from the NCBI database and are included as references. The classification of the specific evolutionary clades is defined. The major amino acid signatures of the virus clades are indicated, including D114N-S202T-S468N for clade 11, V251I-K300E for clade 11.1, S160G-S202T-A214T-S468N for clade 12, H155Q for clade 12.1, and K180I for clade 12.2 of the A(H1N1)pdm09 viruses and Q33R-N145S-N278K for clade TW2010-13 of the A(H3N2) viruses.