Abstract
Early and accurate diagnosis is essential for optimal therapeutic outcomes in patients infected with HIV. Currently, none of the commercially available fourth-generation assays differentiate HIV-1 and HIV-2 antibodies (Ab) or the HIV-1 p24 antigen (Ag). The aim of this study was to evaluate the performance of a novel assay, the BioPlex 2200 HIV Ag-Ab. This assay uses a multiplex flow immunoassay design allowing the simultaneous detection and identification of antibodies to HIV-1 (groups M and O), HIV-2, and the HIV-1 p24 antigen, in addition to providing a traditional composite result. A total of 1,505 routine serum samples were prospectively tested. Results were compared with those from the Architect HIV Combo assay. The sensitivity of the BioPlex 2200 was 100%. The specificity assessed on repeated false-positive samples was 99.5%. In addition, 524 frozen specimens from patients known to be infected with HIV-1 or HIV-2 were tested. Of these specimens, 420 were infected with HIV-1, including 156 of known genotypes, 86 were infected with HIV-2, 7 were infected with HIV-1 and HIV-2, and 11 were from patients with acute HIV infection. Sensitivity was 100% for the HIV genotypes tested. The differentiation capabilities of the BioPlex 2200 HIV Ag-Ab assay for HIV-1, HIV-2, dual HIV-1/HIV-2, and early infections were 100%, 90.7%, 100%, and 90.9%, respectively. The BioPlex 2200 is a sensitive and specific assay that offers advantages over conventional HIV combo assays, also referred to as fourth-generation assays, to accurately differentiate and report HIV-1 p24 antigen and HIV-1 and HIV-2 antibodies.
INTRODUCTION
Early diagnosis is essential for optimal outcomes in patients infected with HIV because it facilitates timely initiation of appropriate care, and it decreases the rate of HIV transmission by 3- to 5-fold (1). The importance of early detection is underlined by studies demonstrating increased life expectancy following early initiation of antiviral treatment. Moreover, several recent high-profile studies have highlighted the potential for limiting viral reservoir expansion and offering protection of innate and specific immunity from the deleterious effects of chronic immune activation by initiating antiretroviral therapy (ART) during acute HIV-1 infection (AHI) (2, 3). For 30 years, remarkable progress has been made in the development of tools for HIV detection. HIV combo assays, also referred to as fourth-generation assays, detect both HIV-1 and HIV-2 antibodies (Ab) and the HIV-1 p24 antigen (Ag) which reduces, compared to third-generation assays, the window period to an average of 2 weeks (4–12).
HIVs display extraordinary genetic diversity due to their fantastic recombination properties. They are subdivided into HIV-1 and -2 and, among HIV-1, 4 groups (M, N, O, and P), of which the pandemic group M includes 9 subtypes and more than 40 circulating recombinant forms (CRFs) as well as numerous unique recombinant forms (URFs). In France, the epidemic in recent years has been characterized by the predominance of subtype B strains but with increases of non-B subtypes (around 50%).
Although the sensitivities and specificities of screening assays have improved, the genetic variability of HIV still represents a challenge, in particular for early detection of infection. For example, a correct serological diagnosis of HIV-2 infection may be missed. The use of HIV-1 Western blot assay as the sole confirmatory test in areas where HIV-2 is not endemic may in fact lead to misclassification of HIV-2-infected individuals as HIV-1 positive. This is due to cross-reactivity between HIV-2 antibodies and envelope glycoproteins of HIV-1. The precise detection of HIV-2 has implications for the choice of antiretroviral treatment (13). Indeed, HIV-2 strains are naturally resistant to nonnucleoside reverse transcriptase inhibitors (NNRTI) and fusion inhibitors and are less sensitive in vitro to some protease inhibitors (14, 15).
Another challenge is posed by HIV-O strains, which are highly divergent from the major group M, leading to their designation as outliers. These strains also display marked intragroup genetic diversity (16). This genetic diversity has important implications for diagnosis and monitoring of HIV-O infection, including risks of false negativity and viral load underestimations (17–19).
New assays allowing the detection and differentiation of HIV-1 (group M and O) and HIV-2 are necessary to improve the diagnosis of HIV infection. Currently, none of the commercially available fourth-generation assays have this capability. The BioPlex 2200 HIV Ag-Ab uses a multiplex flow immunoassay design that permits simultaneous detection, identification, and reporting of antibodies to HIV-1 (groups M and O) and HIV-2 and the HIV-1 p24 antigen in a single reaction vessel.
The aim of this study was to evaluate the sensitivity and specificity of the BioPlex 2200 HIV Ag-Ab assay and its ability to detect and differentiate acute HIV infection (AHI) and HIV-1 and HIV-2 infections.
MATERIALS AND METHODS
Patient samples.
Patient samples were from two populations. The first population consisted of 1,505 fresh specimens obtained during a prospective study between October 2012 and February 2013 from hospitalized patients (Saint-Louis Hospital, Paris, France) for whom screening for HIV infection was requested. The second population included 524 frozen specimens from patients known to be infected by HIV-1 or HIV-2. Of these specimens, 420 were from individuals infected by HIV-1, and 156 of the specimens were of known genotype, including 8 HIV-1 group O samples. There were 86 HIV-2 samples, 7 samples from individuals infected by both HIV-1 and HIV-2, and 11 samples from patients with acute HIV-1 infection (AHI). The study was carried out in accordance with the Declaration of Helsinki. This study was a noninterventional study with no addition to usual procedures. Biological material and clinical data were obtained only for routine viral diagnostic testing according to physicians' directions (no specific sampling and no modification of the sampling protocol). Data analyses were carried out using an anonymized database.
HIV screening assay. (i) BioPlex 2200 HIV Ag-Ab assay.
The BioPlex 2200 HIV Ag-Ab assay uses three reagents. The bead reagent is a mixture of four populations of dyed microparticles, or beads. One dyed bead population is coated with monoclonal antibodies against HIV-1 p24 antigen. Three other dyed bead populations are coated, respectively, with three different antigens: HIV-1 gp160 recombinant protein, a synthetic peptide mimicking HIV-1 group O epitopes, and a peptide mimicking the immunodominant epitope of the HIV-2 envelope protein (BioPlex 2200 HIV Ag-Ab reagent kit, reference 665-3450; Bio-Rad Laboratories, Hercules, CA).
Results for each bead population are determined from a two-point calibration plot from which index values (IDX) are derived. IDX values of <0.90 are nonreactive, values of >1.00 are reactive, and values of 0.90 to 0.99 are in the gray zone. For every sample processed, three internal quality control beads are employed in order to check for detector fluctuations and sample integrity and to normalize the assay signals. An HIV undifferentiated result is yielded when the BioPlex 2200 HIV-Ag-Ab is reactive for both HIV-1 and HIV-2, but with an HIV-2/HIV-1 Ab ratio value that is insufficient to differentiate HIV-1 or HIV-2.
The BioPlex 2200 HIV Ag-Ab assay was designed to have an analytical sensitivity of <12.5 pg/ml for HIV-1 p24 antigen on a panel derived from the National Agency for the Safety of Medicines and Health Products (MSNA) and <2 IU/ml on the WHO HIV international standard NIBSC 60/636. Results for antigen sensitivity are 7.02 pg/ml (range, 6.82 to 7.22 pg/ml) on the MSNA standard and 0.637 IU/ml (range, 0.615 to 0.658 IU/ml) on the WHO standard (BioPlex 2200 HIV Ag-Ab; Bio-Rad Laboratories, Hercules, CA).
(ii) Architect HIV Combo assay.
The Architect HIV Combo assay is a chemiluminescent magnetic microparticle-based immunoassay run on an automated random access instrument. Briefly, the assay is designed to give a single, confounded result based on detection of HIV type 1 antibodies (HIV-1 groups M, O, and N), HIV-2 antibodies, and HIV-1 p24 antigen. Specimens with signal-to-cutoff (S/CO) ratios of 1.0 or greater are considered reactive.
Analysis.
We compared the results of the BioPlex 2200 HIV Ag-Ab assay to those of the Architect HIV Ag/Ab Combo assay according to the manufacturer's instructions (Abbott Diagnostic, Rungis, France). Additional reference tests for HIV detection were performed to discriminate HIV-1 and HIV-2 antibodies, such as the ImmunoComb II HIV-1 & 2 BiSpot differentiation assay (Orgenics, Courbevoie, France) and the New Lav Blot I & II confirmatory assay (Bio-Rad, Marnes la Coquette, France). Also, plasma HIV-RNAs were determined by the Ampliprep/Cobas TaqMan HIV-1 v2.0 assay (Roche Diagnostics, Meylan, France) for RNA-HIV-1 and a noncommercial RNA-HIV-2 reverse transcription (RT)-PCR (20). The sensitivities of the plasma HIV-1- and HIV-2-RNAs were 20 copies/ml and 100 copies/ml, respectively. The viral subtype of HIV-1 was determined with the sequence of the protease and reverse transcriptase (Pol) nucleotide region using a 16-capillary sequencer (ABI PRISM genetic analyzer; Applied Biosystems, Les Ulis, France) along with the Los Alamos HIV sequence database (HIV BLAST [see http://www.hiv.lanl.gov/]).
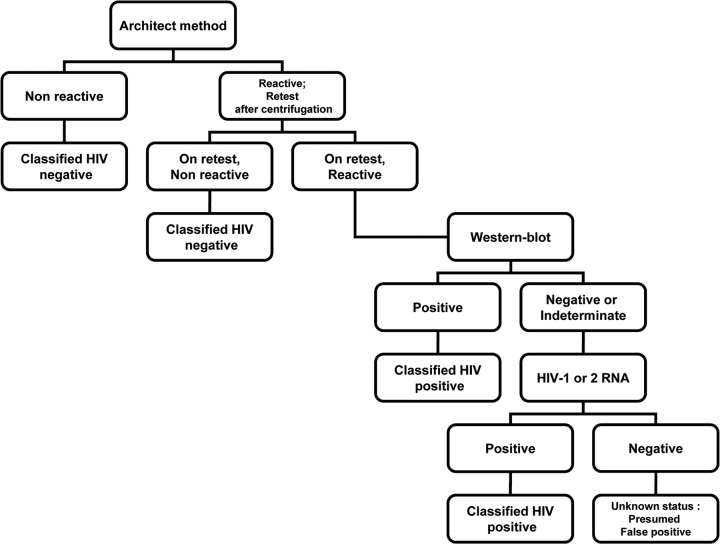
Final classification was established using predicate assays following a test algorithm (Fig. 1). To calculate sensitivity and specificity, specimens were considered to be HIV infected if they were repeatedly reactive by the Architect HIV Ag/Ab Combo assay and reactive by a Western blotting test or if HIV-1 or -2 RNA was detected. They were considered to be AHI if the serology results were indeterminate (using Architect or Western blot) and HIV-1 or -2 RNA was detected. Samples that were discordant on the BioPlex 2200 for final classification were repeated in duplicate after centrifugation.
FIG 1.
Algorithm for determining the final classification of samples by using additional testing.
The BioPlex 2200 HIV Ag-Ab assay (Bio-Rad, Marnes-la-Coquette) results were compared to the Abbott Architect HIV Combo assay results and to final classification results based on additional testing. The sensitivity analysis included HIV-positive samples only, collected during both prospective and retrospective studies. The specificity analysis included only hospitalized patient samples collected during the prospective study.
RESULTS
During the prospective study in March 2013, a total of 1,505 consecutive specimens were tested using the BioPlex 2200 assay, of which 35 were considered HIV infected, based on Architect reactivity and confirmation test results. During the screening, the BioPlex 2200 correctly identified these 35 positive specimens, giving a sensitivity of 100% (95% confidence interval [CI], 96.7 to 100), and 1,461 negative specimens, giving a specificity of 99.4% (95% CI, 99 to 99.8). There were 9 false positives on initial screening. Out of the 9 specimens, repeat testing with the BioPlex 2200 assay reported 1 negative result and 8 repeatedly reactive results, leading to a specificity of 99.5% (95% CI, 99.1 to 99.9%) (Table 1).
TABLE 1.
Specificities and sensitivities of the BioPlex 2200 HIV Ag-Ab assay
| Screening and result | No. of samples that were: |
Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | |
|---|---|---|---|---|
| HIV infected (n = 35) | HIV uninfected (n = 1,470) | |||
| Initial | ||||
| Positive | 35 | 9 | ||
| Negative | 0 | 1,461 | ||
| Performance | 100 (96.7–100) | 99.4 (99.0–99.8) | ||
| Retest | ||||
| Positive | 35 | 8 | ||
| Negative | 0 | 1,462 | ||
| Performance | 100 (96.7–100) | 99.5 (99.1–99.9) | ||
In the retrospective study, a total of 420 HIV-1-positive specimens, 11 AHI specimens (HIV-1), 86 HIV-2-positive specimens, and 7 HIV-1 and HIV-2 specimens from co-infected patients were tested using the BioPlex 2200 assay. All these specimens were reactive on the BioPlex 2200 (Table 2).
TABLE 2.
Samples tested and summary of results
| Sample characterization (by Architect assay, WB, and HIV RNA detection)a | Total no. tested | No. negb | No. positive with BioPlex 2200 HIV Ag-Ab |
Total no. | Sensitivity (%) | Specificity (%) | Diffd capability (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ag p24 | Ab HIV-1 | Ag + Ab | Ab HIV-2 | Undiffc | |||||||
| Negative | 1,470 | 1,462 | 1 | 7 | 0 | 0 | 0 | 8 | 99.5 | ||
| HIV-1 (no AHI) | 420 | 0 | 0 | 420 | 0 | 0 | 420 | 100 | 100 | ||
| HIV-1 (AHI) | 11 | 0 | 2 | 3 | 5 | 0 | 1 | 11 | 100 | 90.9 | |
| HIV-2 | 86 | 0 | 0 | 0 | 0 | 78 | 8 | 86 | 100 | 90.7 | |
| HIV-1 and HIV-2 | 7 | 0 | 0 | 0 | 0 | 0 | 7 | 7 | 100 | 100 | |
WB, Western blot.
neg, negative.
Undiff, undifferentiating.
Diff, differentiation.
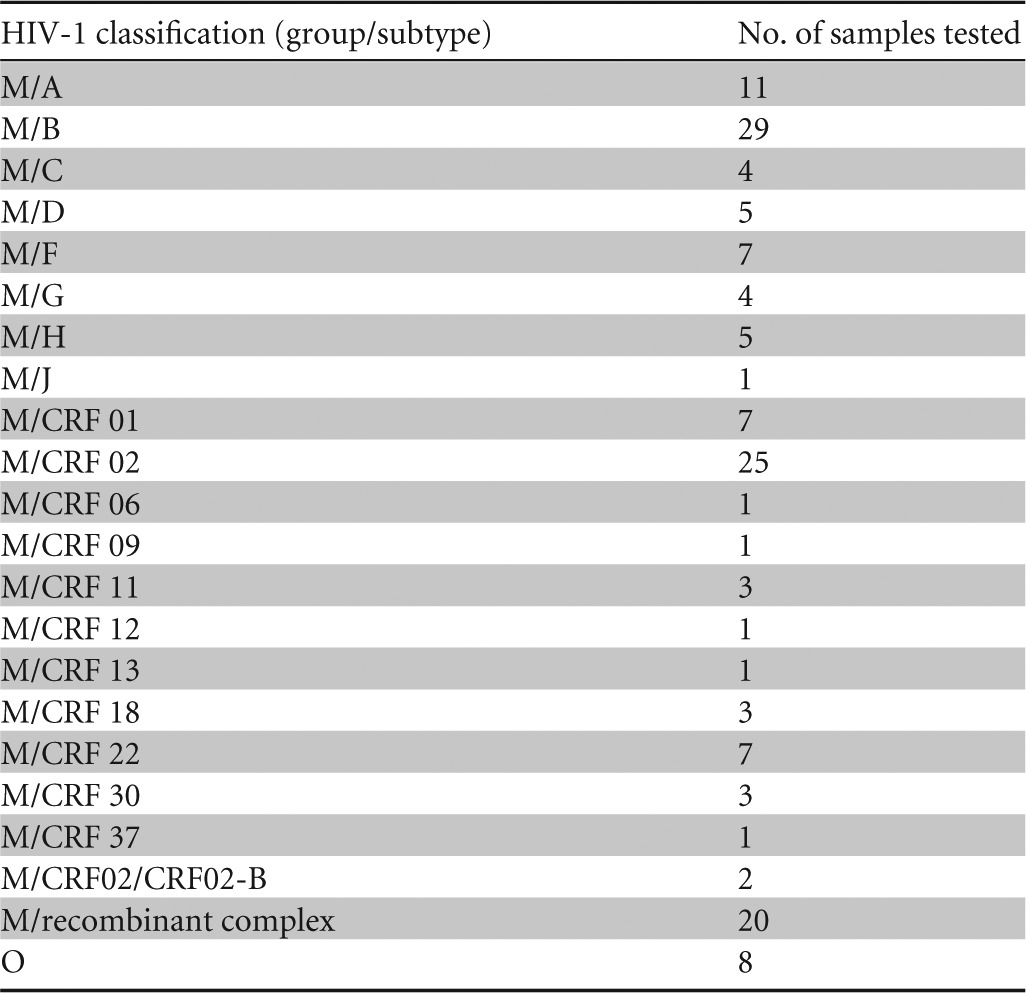
The 420 specimens from HIV-1-infected-patients tested reactive on the BioPlex 2200 HIV Ag-Ab, with only HIV-1 Ab reactivity. At least 28 different genotypes were represented, and the sensitivity was 100% (95% CI, 99.3 to 100) (Table 3). All specimens were correctly classified as HIV-1 reactive by the BioPlex 2200, indicating a differentiation capability of 100% (95% CI, 99.3 to 100). The BioPlex 2200 assay's sensitivity was unaffected by the HIV-1 group or subtype. Eight HIV-1 group O samples were classified as HIV-1.
TABLE 3.
HIV-1 genotypes tested on the BioPlex 2200 HIV Ag-Ab assay
All of the 11 AHI HIV-1 specimens tested reactive on the BioPlex 2200. Five of them were reactive with both HIV-1 Ab and HIV-1 p24 Ag, two with only HIV-1 p24 Ag, two with only HIV-1 Ab, and one with an HIV undifferentiated result.
All of the 86 specimens considered HIV-2 infected tested reactive on the BioPlex 2200, with 78 specimens giving an HIV-2 Ab-reactive result, and 8 giving an HIV-undifferentiated result. These results gave a sensitivity for HIV-2-positive specimens of 100% (95% CI, 96.6 to 100) and an HIV-1/2 differentiation capability of 90.7% (95% CI, 82.5 to 95.9).
All of the specimens positive for both HIV-1 and HIV-2 tested reactive on the BioPlex 2200, with HIV-undifferentiated results giving a sensitivity of 100% for these specimens.
DISCUSSION
The results of this study show that the BioPlex 2200 is highly sensitive and specific in clinical sample populations and has performance comparable to that of the Architect HIV Combo assay. In contrast to the alternative assays, the BioPlex 2200 assay offers the advantage of calculating and reporting separate values for HIV-1 p24 Ag and HIV-1 (group M and O) and HIV-2 antibodies. Currently, to our knowledge, no other commercial screening assay can differentiate HIV-1 (group M and O) and HIV-2. Individual reporting of HIV-1 group M and O results are for research use only (RUO) since the BioPlex 2200 will not report results for the two bead populations separately.
The detection of AHI has been shown to be beneficial for the prompt initiation of appropriate antiretroviral therapy. In fact, HIV primary infections are now considered a therapeutic emergency to limit, by early and adapted highly active antiretroviral therapy (HAART), the viral reservoir constitution. In this way, the BioPlex 2200 is an interesting test because it allows differentiation between HIV-1 p24 antigen and HIV antibody reactivity. During this study, the BioPlex 2200 showed excellent sensitivity for the detection of AHI by detecting HIV-1 p24 antigen reactivity in 7 of the 11 AHI samples.
Because of the growing number of people with HIV-1 group M non-B-subtype infections, it is essential that HIV assays detect infection independently of subtype. Studies have demonstrated that some fourth-generation HIV assays do not detect certain HIV-1 group M, non-B subtypes, yielding false-negative results (8, 21). The BioPlex 2200 HIV Ag-Ab assay had 100% sensitivity for all of the HIV-1 groups (M and O) and subtypes (n > 20) tested and for HIV-2. Various HIV-1 subtypes tested in our study were represented as major recombinant forms and very divergent strains currently observed in Africa, Europe, and elsewhere.
Two samples from individuals infected by HIV-1 group O tested positive on the BioPlex 2200 with HIV-O antibody reactivity with no cross-reactivity with the group M antigen target. This shows the importance of including BioPlex 2200 HIV Ag-Ab beads with the HIV-1 group O-specific epitope. This observation suggests that it may be possible for specialists to observe, under RUO conditions, specific binding reactivity to characterized samples suspected of being group O positive.
The BioPlex 2200 HIV Ag-Ab assay identifies patients infected by HIV-1 or HIV-2 and has excellent specificity to discriminate between two HIV infections, allowing early initiation of adapted antiretroviral treatment and avoiding inappropriate therapy in HIV-2 patients with natural resistance to NNRTI.
It is conventional to classify the HIV screening enzyme immunoassays (EIAs) according to their technological advancement. So we went past the first generation to the fourth generation according to the nature of the antigens, the EIA detection formats, and the detection of p24 antigen. The shift to a test using a sandwich method with detection and differentiation by the Luminex technology allows us to consider this new tool a new generation of test.
This study shows that the next-generation BioPlex 2200 HIV Ag-Ab assay performs well in the diagnosis of HIV infection, with excellent sensitivity and specificity. In conclusion, the BioPlex 2200 could be an alternative to fourth-generation assays by allowing screening for early infection for rapid HAART initiation and simultaneous differentiation of HIV-1 and HIV-2.
ACKNOWLEDGMENTS
We are grateful to Marie-Charlotte Bassara for her technical assistance.
We thank Bio-Rad Laboratories for their financial support.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, Hakim JG, Kumwenda J, Grinsztejn B, Pilotto JHS, Godbole SV, Mehendale S, Chariyalertsak S, Santos BR, Mayer KH, Hoffman IF, Eshleman SH, Piwowar-Manning E, Wang L, Makhema J, Mills LA, de Bruyn G, Sanne I, Eron J, Gallant J, Havlir D, Swindells S, Ribaudo H, Elharrar V, Burns D, Taha TE, Nielsen-Saines K, Celentano D, Essex M, Fleming TR, HPTN 052 Study Team 2011. Prevention of HIV-1 infection with early antiretroviral therapy. N. Engl. J. Med. 365:493–505. 10.1056/NEJMoa1105243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ananworanich J, Schuetz A, Vandergeeten C, Sereti I, de Souza M, Rerknimitr R, Dewar R, Marovich M, van Griensven F, Sekaly R, Pinyakorn S, Phanuphak N, Trichavaroj R, Rutvisuttinunt W, Chomchey N, Paris R, Peel S, Valcour V, Maldarelli F, Chomont N, Michael N, Phanuphak P, Kim JH, RV254/SEARCH 010 Study Group 2012. Impact of multitargeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One 7:e33948. 10.1371/journal.pone.0033948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sáez-Cirión A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard J-P, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, Study Group ANRS VISCONTI 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. Plos Pathog. 9:e1003211. 10.1371/journal.ppat.1003211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber B, Orazi B, Raineri A, Thorstensson R, Bürgisser P, Mühlbacher A, Areal C, Eiras A, Villaescusa R, Camacho R, Diogo I, Roth HJ, Zahn I, Bartel J, Bossi V, Piro F, Atamasirikul K, Permpikul P, Webber L, Singh S. 2006. Multicenter evaluation of a new 4th generation HIV screening assay Elecsys HIV combi. Clin. Lab. 52:463–473. 10.1007/s00430-012-0250-5 [DOI] [PubMed] [Google Scholar]
- 5.Weber B, Fall EH, Berger A, Doerr HW. 1998. Reduction of diagnostic window by new fourth-generation human immunodeficiency virus screening assays. J. Clin. Microbiol. 36:2235–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malm K, von Sydow M, Andersson S. 2009. Performance of three automated fourth-generation combined HIV antigen/antibody assays in large-scale screening of blood donors and clinical samples. Transfus. Med. Oxf. Engl. 19:78–88. 10.1111/j.1365-3148.2009.00907.x [DOI] [PubMed] [Google Scholar]
- 7.Gürtler L, Mühlbacher A, Michl U, Hofmann H, Paggi GG, Bossi V, Thorstensson R, G-Villaescusa R, Eiras A, Hernandez JM, Melchior W, Donie F, Weber B. 1998. Reduction of the diagnostic window with a new combined p24 antigen and human immunodeficiency virus antibody screening assay. J. Virol. Methods 75:27–38. 10.1016/S0166-0934(98)00094-9 [DOI] [PubMed] [Google Scholar]
- 8.Ly TD, Martin L, Daghfal D, Sandridge A, West D, Bristow R, Chalouas L, Qiu X, Lou SC, Hunt JC, Schochetman G, Devare SG. 2001. Seven human immunodeficiency virus (HIV) antigen-antibody combination assays: evaluation of HIV seroconversion sensitivity and subtype detection. J. Clin. Microbiol. 39:3122–3128. 10.1128/JCM.39.9.3122-3128.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brust S, Duttmann H, Feldner J, Gürtler L, Thorstensson R, Simon F. 2000. Shortening of the diagnostic window with a new combined HIV p24 antigen and anti-HIV-1/2/O screening test. J. Virol. Methods 90:153–165. 10.1016/S0166-0934(00)00229-9 [DOI] [PubMed] [Google Scholar]
- 10.Saville RD, Constantine NT, Cleghorn FR, Jack N, Bartholomew C, Edwards J, Gomez P, Blattner WA. 2001. Fourth-generation enzyme-linked immunosorbent assay for the simultaneous detection of human immunodeficiency virus antigen and antibody. J. Clin. Microbiol. 39:2518–2524. 10.1128/JCM.39.7.2518-2524.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Binsbergen J, Siebelink A, Jacobs A, Keur W, Bruynis F, van de Graaf M, van der Heijden J, Kambel D, Toonen J. 1999. Improved performance of seroconversion with a 4th generation HIV antigen/antibody assay. J. Virol. Methods 82:77–84. 10.1016/S0166-0934(99)00086-5 [DOI] [PubMed] [Google Scholar]
- 12.Van Binsbergen J, Keur W, Siebelink A, van de Graaf M, Jacobs A, de Rijk D, Nijholt L, Toonen J, Gürtler LG. 1998. Strongly enhanced sensitivity of a direct anti-HIV-1/-2 assay in seroconversion by incorporation of HIV p24 ag detection: a new generation Vironostika HIV Uni-Form II. J. Virol. Methods 76:59–71. 10.1016/S0166-0934(98)00126-8 [DOI] [PubMed] [Google Scholar]
- 13.Chiara M, Rony Z, Homa M, Bhanumati V, Ladomirska J, Manzi M, Wilson N, Alaka D, Harries AD. 2010. Characteristics, immunological response & treatment outcomes of HIV-2 compared with HIV-1 & dual infections (HIV 1/2) in Mumbai. Indian J. Med. Res. 132:683–689 [PMC free article] [PubMed] [Google Scholar]
- 14.Damond F, Brun-Vezinet F, Matheron S, Peytavin G, Campa P, Pueyo S, Mammano F, Lastere S, Farfara I, Simon F, Chene G, Descamps D. 2005. Polymorphism of the human immunodeficiency virus type 2 (HIV-2) protease gene and selection of drug resistance mutations in HIV-2-infected patients treated with protease inhibitors. J. Clin. Microbiol. 43:484–487. 10.1128/JCM.43.1.484-487.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves JD, Doms RW. 2002. Human immunodeficiency virus type 2. J. Gen. Virol. 83:1253–1265 [DOI] [PubMed] [Google Scholar]
- 16.Roques P, Robertson DL, Souquière S, Damond F, Ayouba A, Farfara I, Depienne C, Nerrienet E, Dormont D, Brun-Vézinet F, Simon F, Mauclère P. 2002. Phylogenetic analysis of 49 newly derived HIV-1 group O strains: high viral diversity but no group M-like subtype structure. Virology 302:259–273. 10.1006/viro.2002.1430 [DOI] [PubMed] [Google Scholar]
- 17.Depatureaux A, Leoz M, De Oliveira F, Gueudin M, Damond F, Descamps D, Brun-Vézinet F, Lemée V, Simon F, Barin F, Plantier J-C. 2010. Specific diagnosis and follow-up of HIV-1 group O infection: RES-O data. Med. Mal. Infect. 40:669–676 (In French.) 10.1016/j.medmal.2010.04.011 [DOI] [PubMed] [Google Scholar]
- 18.Gueudin M, Plantier JC, Lemée V, Schmitt MP, Chartier L, Bourlet T, Ruffault A, Damond F, Vray M, Simon F. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500–505. 10.1097/QAI.0b013e31803260df [DOI] [PubMed] [Google Scholar]
- 19.Plantier J-C, Djemai M, Lemée V, Reggiani A, Leoz M, Burc L, Vessière A, Rousset D, Poveda J-D, Henquell C, Gautheret-Dejean A, Barin F. 2009. Census and analysis of persistent false-negative results in serological diagnosis of human immunodeficiency virus type 1 group O infections. J. Clin. Microbiol. 47:2906–2911. 10.1128/JCM.00602-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damond F, Collin G, Descamps D, Matheron S, Pueyo S, Taieb A, Campa P, Benard A, Chêne G, Brun-Vezinet F. 2005. Improved sensitivity of human immunodeficiency virus type 2 subtype B plasma viral load assay. J. Clin. Microbiol. 43:4234–4236. 10.1128/JCM.43.8.4234-4236.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudy C, Moreau A, Brunet S, Descamps J-M, Deleplanque P, Brand D, Barin F. 2004. Subtype B human immunodeficiency virus (HIV) type 1 mutant that escapes detection in a fourth-generation immunoassay for HIV infection. J. Clin. Microbiol. 42:2847–2849. 10.1128/JCM.42.6.2847-2849.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]