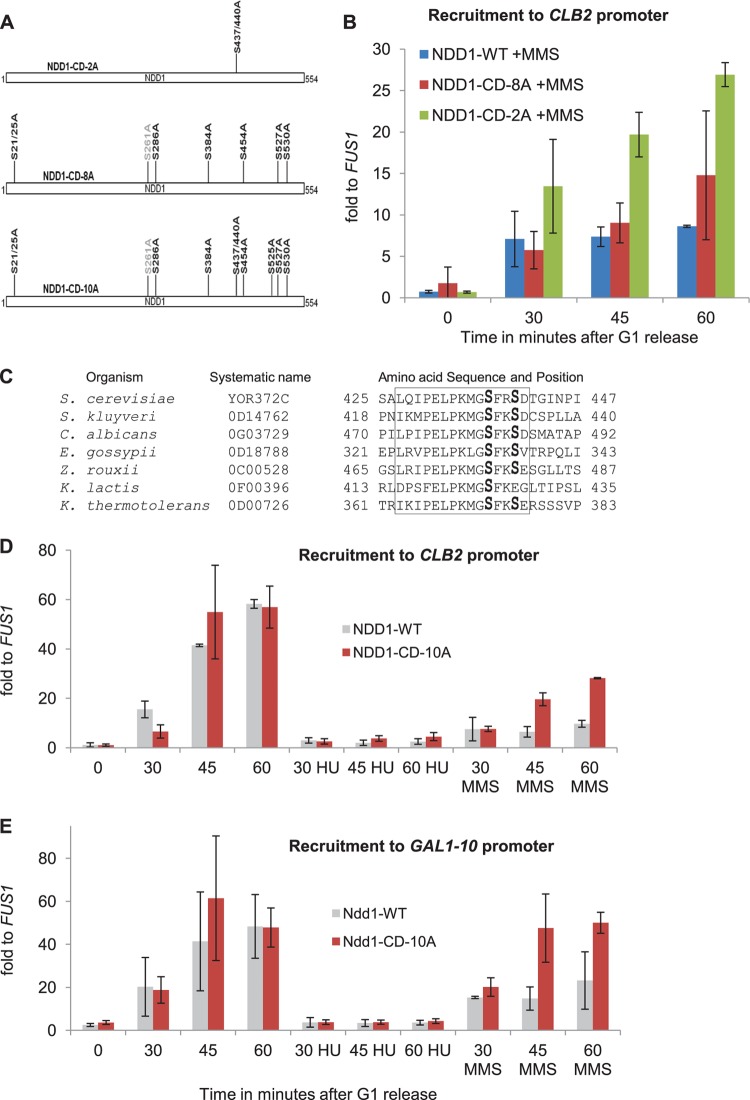
FIG 7.
MMS-dependent phosphorylations of Ndd1 prevent its recruitment to the CLB2 promoter as well as its interaction with the FHA domain of Fkh2. (A) Model showing a series of Ndd1 checkpoint-defective mutants (NDD1-CD) used in the experiments whose results are presented in panels B, D, and E. (B) Conditional GAL promoter-driven NDD1 strains (based on strain JV323) expressing either NDD1-WT–HA, NDD1-CD-2A–HA, or NDD1-CD-8A–HA from endogenous promoters were synchronized in G1 and released into YEP-glucose with MMS. Ndd1 recruitment to the CLB2 promoter was examined by ChIP, followed by qPCR analysis. Error bars indicate standard deviations in three independent experiments. (C) Pairwise sequence alignment of various Ndd1 homologues indicates that S437 and S440 and the surrounding amino acids in the yeast S. cerevisiae Ndd1 are conserved in other yeast species. S. kluyveri, Saccharomyces kluyveri; E. gossypii, Eremothecium gossypii; C. albicans, Candida albicans; Z. rouxii, Zygosaccharomyces rouxii; K. lactis, Kluyveromyces lactis; K. thermotolerans, Kluyveromyces thermotolerans. (D and E) Recruitment of Ndd1-CD-10A mutant to the CLB2 promoter (D) as well as its FHA domain-mediated recruitment to the GAL1-10 promoter (E) is significantly reestablished upon MMS treatment but is still abolished in HU-treated cells. A conditional GAL promoter-driven NDD1 strain (JV323) expressing either NDD1-WT-HA and GAL4DBD-fkh21-306 or NDD1-CD-10A–HA and GAL4DBD-fkh21-306 was synchronized in G1 and released into YEP-glucose with or without MMS or HU. Ndd1 recruitment to the CLB2 and GAL1-10 promoters was examined by ChIP, followed by qPCR analysis. The data sets used for Ndd1-WT releases were the same as those used for the experiment whose results are shown in Fig. 2F. Error bars indicate standard deviations in at least two independent experiments.