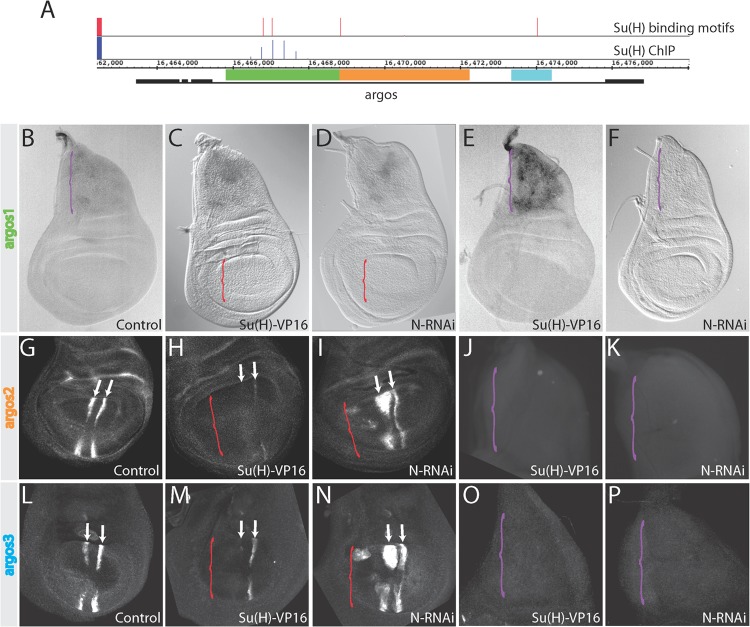
FIG 2.
Distinct Notch regulation is mediated by separable enhancers. (A) Schematic of the argos gene. Black boxes indicate exons and black lines indicate introns. Red bars, conserved Su(H) binding motifs where the height of the bar indicates the patser score (5.1 to 9.8). Blue bars, Su(H)-bound regions identified by ChIP in DmD8 cells (0.3- to 1.4-fold enrichment on a log2 scale). argos1, argos2, and argos3 enhancer regions are indicated by the green, orange, and blue rectangles, respectively. (B to P) Expression from the indicated enhancers in wing discs under different conditions: argos1 (B to F), argos2 (G to K), or argos3 (L to P) reporters in wild-type (B, G, and L), Notch pathway activation [Su(H)-VP16] (C, E, H, J, M, and O), or Notch suppression (N-RNAi) (D, F, I, K, N, and P) conditions. Brackets indicate regions where Notch pathway activity was manipulated: panels C, D, H, I, M, and N, wing pouch expression using salpE80-Gal4 (red brackets); panels E, F, J, K, O, and P, AMP expression using 1151-Gal4 (purple bracket). In panels B to F, whole discs with both thorax (AMPs) and wing pouch are shown; in panels G to I and L to N, wing pouch only is shown; in panels J to K and O to P, thorax (AMPs) only is shown. Arrows mark the L3 and L4 veins.