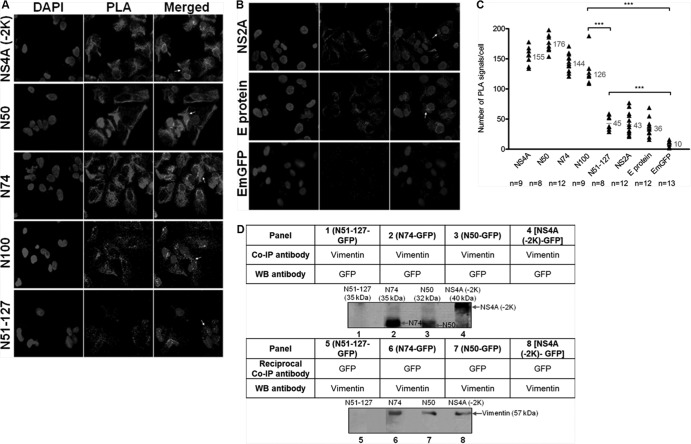
FIG 6.
N50 is the specific interacting region of DENV NS4A with vimentin. In situ PLA exhibits interaction of vimentin with NS4A-EmGFP fusion proteins. (A) Immunofluorescence detection of PLA signals (white arrows) in transfected cells stably expressing different regions of NS4A. Vimentin- and GFP-specific primary antibodies were used. The number of in situ PLA signals represents the vimentin-NS4A fusion protein (region of interest) interactions, and the nuclei were stained with DAPI. Individual events of PLA signal were used as reading output and quantified. (B) Plasmids expressing NS2A protein, E protein, and GFP alone were used as controls. The images were taken at a magnification of ×100. (C) Quantification of PLA-positive foci detected in stably transfected Huh-7 cells. In situ PLA signals per cell were quantitated using the software program CellProfiler (17) and ImageJ program by spot counting (18).The numbers of cells counted (n) are shown, and the average values are shown in gray. Statistical analyses were carried out using one-way ANOVA and Bonferroni post hoc test (GraphPad Software). Values that are significantly different (P < 0.001) from each other are indicated by a bar and three asterisks. (D) Co-IP of vimentin with different NS4A-GFP recombinant fusion proteins. Total cell lysates were incubated with vimentin or GFP antibodies, and Dynabeads protein G were added to capture the protein complex. Samples with eluted target proteins were analyzed by Western blotting (WB) using anti-GFP or antivimentin antibodies. Panels 1 to 4 represent Co-IP procedure, and panels 5 to 8 represent reciprocal Co-IP. The specific protein bands representing different truncated NS4A-GFP fusion proteins and the bands representing vimentin are indicated by the arrows.