FIG 6.
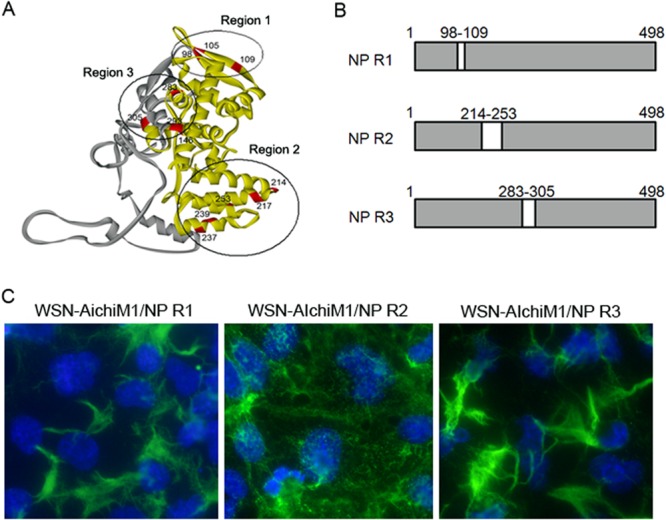
The region of Aichi NP required to inhibit filament formation by the WSN-AichiM1 virus. (A) Crystal structure of NP highlighting the locations of residues that differ between the WSN and Aichi viruses. The N-terminal 305 residues and the rest of the regions are shown in yellow and gray, respectively. Region 1 (R1), R2, and R3 are circled. (B) Schematic diagram of chimeric NP proteins. The amino acid residues of WSN NP located in R1, R2, or R3 were replaced with the corresponding Aichi NP residues to create NP R1, NP R2, and NP R3 chimeras. (C) MDCK cells were infected with recombinant WSN-AichiM1 viruses expressing the R1, R2, or R3 NP chimera at an MOI of 1, as indicated. Cell surfaces were stained for viral proteins with anti-H1N1 goat serum followed by Alexa Fluor 488 anti-goat IgG.
