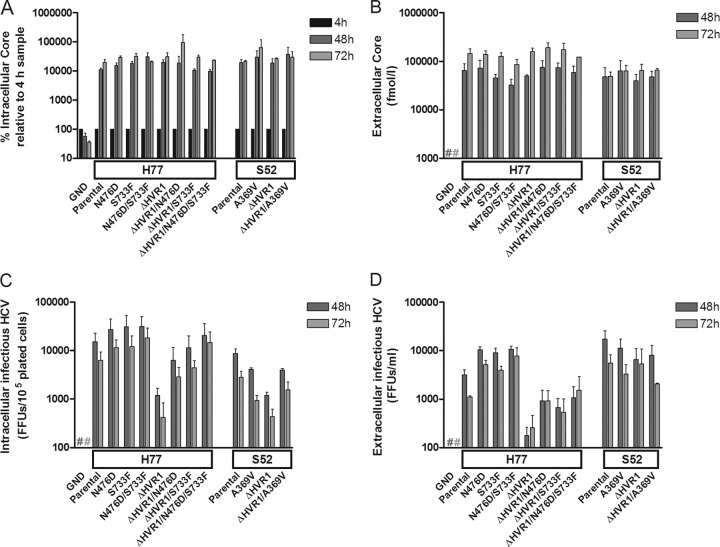
FIG 1.
HCV RNA transfection of S29 cells with low CD81 expression suggested that HVR1 deletion and HVR1-related adaptive envelope mutations had no effect on HCV replication/translation and assembly/release. The boxes on the x axes indicate which Core-NS2 JFH1-based recombinant served as the backbone for the indicated mutations, and Parental signifies the unmodified recombinant. A J6/JFH1 recombinant with the replication-deficient GND mutation was included as a negative control. Cells were plated and transfected with the indicated HCV RNA as described in Materials and Methods to allow quantification of intra- and extracellular Core at 4 h, 48 h, and 72 h posttransfection, as well as intra- and extracellular infectivity titers. (A) Protein was harvested from the S29 cells at 4 h, 48 h, and 72 h posttransfection and analyzed by Core ELISA. The Core values were normalized to the 4-h value, representing transfection efficiency. (B) S29 cell culture supernatants were collected at 48 h and 72 h posttransfection. Core values were measured directly on the supernatants by Core ELISA. (C) S29 cells were subjected to freeze-thaw cycles and cleared of cellular debris by centrifugation prior to HCV infectivity titration in triplicate to obtain intracellular infectious particles at 48 h and 72 h posttransfection. (D) S29 cell culture supernatants were HCV infectivity titrated in triplicate. The entire single-cycle production assay was repeated, and the values represent the means of the two independent experiments performed as described above, with error bars representing standard errors of the mean (SEM). #, values below the cutoff.