FIG 3.
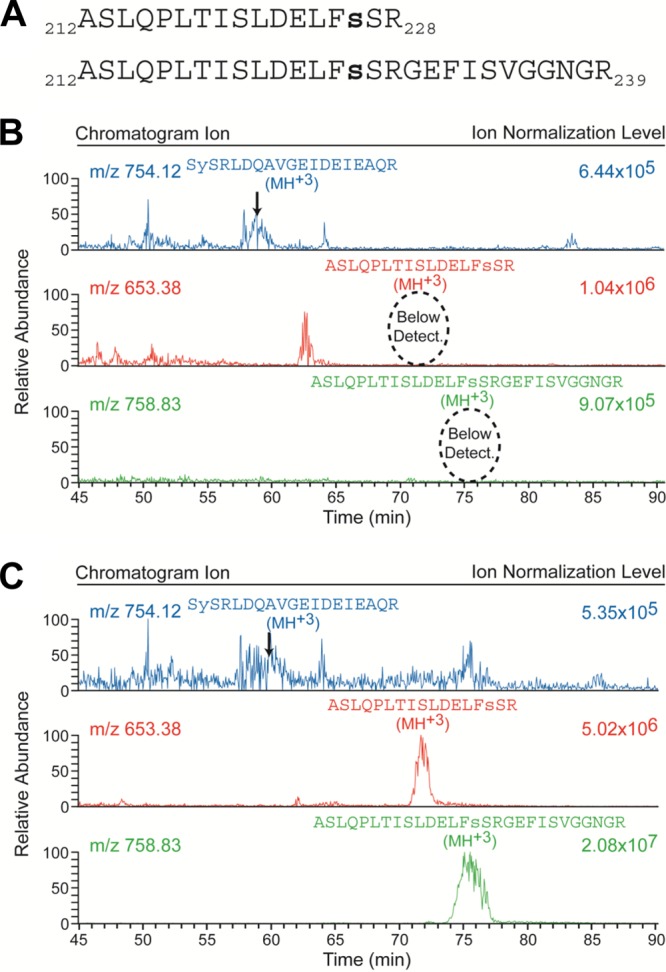
Phosphorylation at Ser226 and Ser227 is below the limit of detection. (A) Sequences of the two synthetic phosphopeptides representing the tryptic (residues 212 through 228) and extended (residues 212 through 239 with missed cleavage at R228) peptides of the VSV P protein that encompass the putative Ser226 and Ser227 phosphorylation sites. Both of these synthetic peptides were phosphorylated at residue Ser226, as indicated by the small bold letter s. (B) Chromatograms obtained from a biological sample without the addition of the two synthetic phosphopeptides. The top chromatogram presents the m/z = 754.12 ion that represents the +3 charge state of the newly identified Ser13/Tyr14 phosphopeptide; the indicated elution peak was verified via inspection of its corresponding MS2 CID spectrum as illustrated in Fig. 2B. The bottom two chromatograms present the m/z = 653.38 and 758.83 ions that represent the +3 charge state of the tryptic and extended synthetic phosphopeptides, respectively (as displayed in panel A), occurring in the biological sample. (C) The corresponding chromatograms obtained from a biological replicate of the sample represented in panel B but with 2 pmol of each of the two synthetic phosphopeptides in panel A added. A comparison of the corresponding bottom two chromatograms in panels B and C reveals that phosphorylated Ser226 and Ser227 are below the limit of detection in the biological sample and thus exist in, at most, <1% of the total abundance of P protein. Note that the individual chromatograms are each scaled independently by the indicated ion normalization level.
