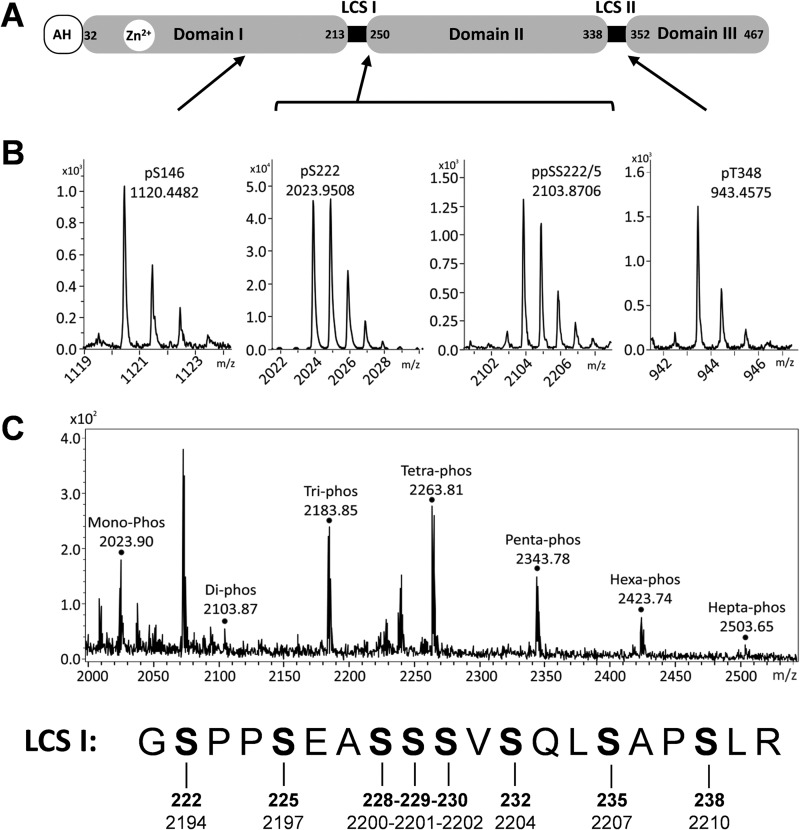
FIG 1.
Mass spectrometric analysis of NS5A phosphorylation. (A) Schematic of NS5A structure showing amino acid residue numbers for JFH-1. AH, amphipathic helix; LCS: low-complexity sequence. (B) Mass spectra of NS5A phosphopeptides after analysis by high-performance liquid chromatography (HPLC)-MS/MS. The main monoisotopic peaks were selected for collision-induced dissociation (MS/MS) to allow the precise assignment of peptide and the phosphorylated residues (see Fig. S1 to S4 in the supplemental material). The locations of the four phosphopeptide species within NS5A are indicated. (C) The mass spectra of the NS5A phosphopeptides corresponding to the highly phosphorylated LCS I region, coeluted from the HPLC, resulting in simultaneous observation of all species. Peaks identified as mono- to heptaphosphorylated species are indicated. The sequence of this peptide, together with the amino acid sequence numbering for JFH-1 NS5A (bold) and the JFH-1 polypeptide, is presented below. Monoisotopic peaks for the tri, tetra, and penta species were selected for MS/MS; peptide sequence could be assigned, but exact mapping of phosphorylated residues was hindered by the presence of multiple isomers of each species. Residues S229, S230, S232, and S235, however, were observed as phosphorylated but in unassignable conformations. No MS/MS data could be acquired for the hexa- or heptaphosphorylated species.