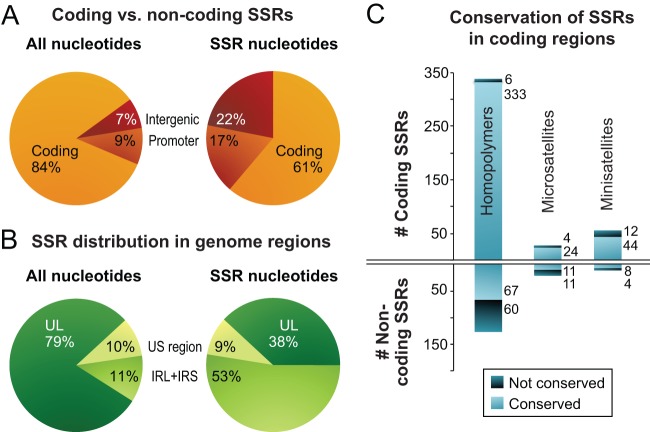
FIG 4.
Localization and conservation of SSRs in HSV-1 strains. (A) SSRs in reference strain 17 are overrepresented in noncoding regions. The pie chart on the left shows the distribution of all nucleotides in the trimmed genome alignment among protein-coding and noncoding (promoter and intergenic) regions. The pie chart on the right shows the distribution of SSR-encoding nucleotides among protein-coding and noncoding regions. (B) SSRs in reference strain 17 are more common in the large repeat regions. The pie chart on the left shows the distribution of all nucleotide bases in the trimmed genome (Fig. 2B) among the unique long (UL), unique short (US), and internal repeat (IRL+IRS) regions. The pie chart on the right shows the distribution of SSR bases in UL, US, and IRL+IRS. (C) Although protein-coding SSRs outnumber noncoding repeats, they are more likely to be conserved in length. An SSR was counted as conserved if it had the same position and length (same number of repeated units) in a majority of strains. Coding SSRs are largely conserved in length (pale blue versus dark blue); the number of SSRs in each group is shown to the right of the histograms. In contrast, there are approximately equivalent numbers of conserved versus nonconserved SSRs in noncoding regions. SSRs with incomplete sequences in more than half the strains (gray in Fig. 3) were excluded.