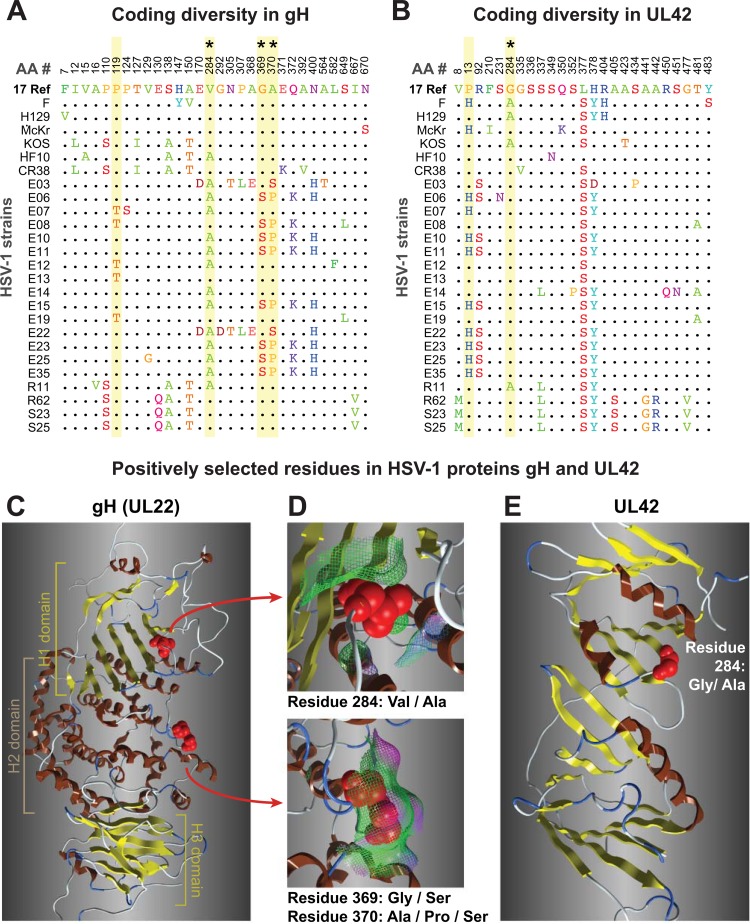
FIG 7.
Coding diversity and positive selection of residues in the HSV-1 entry protein gH and the DNA-binding protein UL42. (A and B) Amino acid sequence alignments of gH (UL22) (A) and UL42 (B) from 26 HSV-1 strains, showing only those positions where a residue varies in one or more strains compared to the residue in reference strain 17 (top line). Positions in the sequence are shown along the top. Yellow shading denotes residues exhibiting positive selection (Codeml, P > 99%; see Materials and Methods for details), and asterisks (*) mark those visible as red spheres on the 3-dimensional models in the panels below. (B) UL42 alignment from 26 HSV-1 strains, in which positive selection was detected for 829 residue 13 (P > 99%) and 284 (P > 95%). (C) Ribbon diagram of the HSV-1 gH ectodomain, homology modeled using the crystal structure of HSV-2 gH (77). Highlighted residues fall into the H1 and H2 domains described previously (77). (D) Surface interactions (surface indicated by mesh, color coded as follows: green, hydrophobic; pink, H bonding; blue, polar) are low for residue 284 (top) and are greater for the exposed pair of positively selected residues 369 and 370. (E) The available structure of UL42 (153) captures only residue 284, which lies adjacent to residues proposed to interact with DNA and with HSV-1 DNA polymerase (Pol [UL30]).