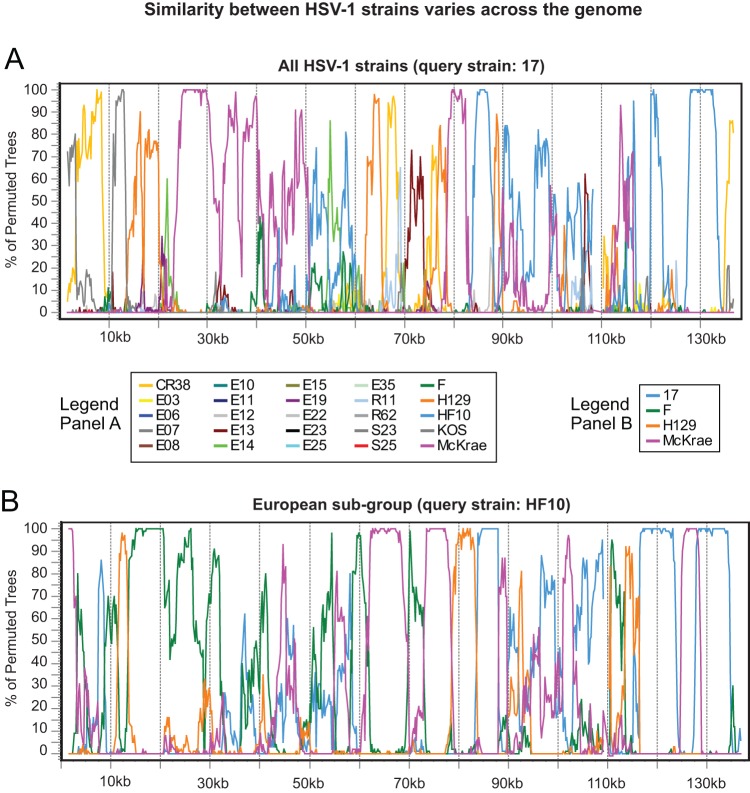
FIG 9.
Bootscan analyses of similarity between HSV-1 strains contain breakpoints suggesting frequent recombination. (A) Similarity plot of HSV-1 reference strain 17 versus all other strains, demonstrating that recombination occurs throughout the tree. The longest colinear area of similarity (between strains 17 and McKrae; rose line) is about 30 kb. The trimmed format of the HSV-1 strain 17 genome (Fig. 1B) was used as the query sequence. (B) Similarity plot of the European subgroup (as shown in Fig. 8) of the HSV-1 collection, with HF10 used as the query sequence. There is extensive recombination even within genetically similar geographical clusters. The longest colinear area of similarity (between HF10 and F; green line) is about 20 kb. Bootscan parameters were as follows: 3-kb window, 200-bp step size, GapStrip on, 100 repetitions, Kimura (2 parameter), T/t = 2.0, neighbor joining.