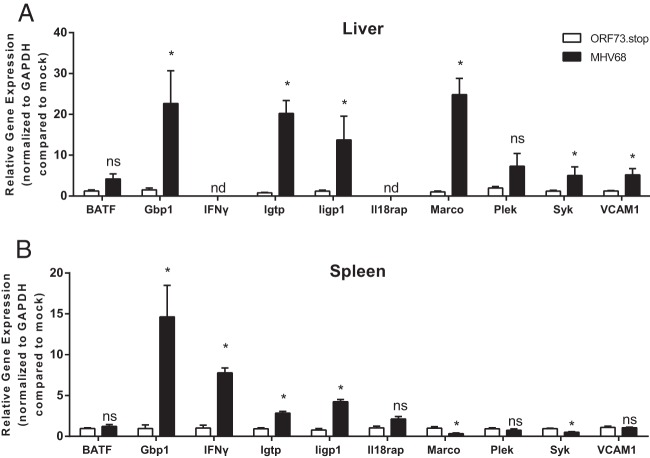
FIG 2.
Validation of microarray analyses by qRT-PCR. RNA pooled from three to four mice per condition was treated with Turbo DNase (Life Technologies) before cDNA synthesis using 1 μg RNA, oligo(dT)12-18, and Superscript II reverse transcriptase (Life Technologies). qPCR was performed with cDNA from livers (A) or spleens (B) using Power SYBR green master mix (Applied Biosystems) with the indicated gene-specific primers (Table 1). Data were collected from 4 independent experiments. Transcript levels were normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) within each sample and compared to the level in mock-infected mice using the ΔΔCT method (where CT is the threshold cycle). Data are presented as means ± SEM. Statistical analyses were performed by using the Mann-Whitney U test. ∗, P < 0.05; ns, not significant; nd, could not reliably detect target.