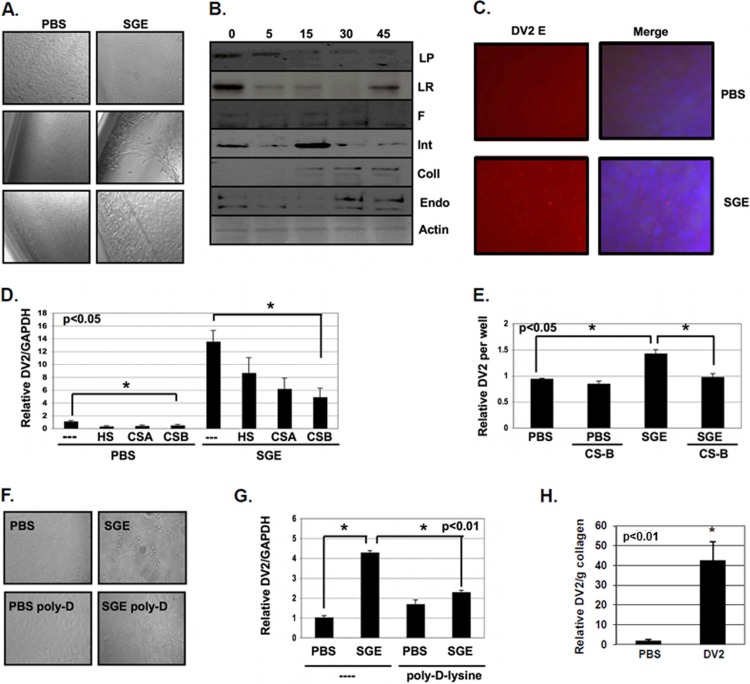
FIG 3.
SGE enhances virus attachment to exposed glycosaminoglycans and induces cell migration. (A) NIH 3T3 MEFs were treated with buffer alone (PBS) or with 1.0 SGE for 10 min and then allowed to incubate for 4 h at 37°C before observation by phase-contrast light microscopy. (B) NIH 3T3 MEFs were treated with 1.0 SGE for 0, 5, 15, 30, and 45 min. Equal volumes of protein lysates were loaded onto SDS-PAGE gels, protein was transferred to PVDF membranes, and the expression of multiple proteins was analyzed by Western blotting. Laminin protein (LP), laminin receptor (LR), fibronectin (F), integrin (Int), collagen type I (Coll), endo180 (Endo), and actin were used. (C) NIH 3T3 MEFs were treated with buffer alone (PBS) or with 2.5 SGE for 10 min and inoculated with DENV2 at an MOI of 1 for 1 h. Unbound virus was washed from cells, and cells were fixed with paraformaldehyde and stained for DENV2 envelope (red) and DAPI (blue). (D) DENV2 was untreated (−) or pretreated with 100 μg/ml heparan sulfate (HS), chondroitin sulfate A (CS-A), and chondroitin sulfate B (CS-B) for 30 min at room temperature. NIH 3T3 MEFs were then treated with buffer alone (PBS) or with 1.0 SGE for 10 min and then inoculated with glycosaminoglycan-treated or untreated DENV2 at an MOI of 1 for 1 h at 37°C. Unbound virus was removed, and then cells were allowed to incubate for 18 h. Total RNA was extracted, and DENV2 vRNA and GAPDH expression was analyzed by RT-qPCR. DENV2 vRNA was normalized to GAPDH. (E) ECM material was isolated on cell culture plates by washing off monolayers of NIH 3T3 MEF cells with 10 mM EDTA solution in PBS for 10 min at 37°C. Wells were either treated with buffer alone (PBS) or treated with 2.0 SGE for 10 min at 37°C. Wells were then inoculated with untreated or CS-B-treated DENV2 for 1 h at 37°C. Unbound virus was removed, total RNA was extracted, and DENV2 vRNA expression was analyzed by RT-qPCR and normalized per well. (F and G) MEFs were seeded onto either untreated (−) or poly-d-lysine-treated (poly-d-lysine) polystyrene cell culture plates and treated with buffer alone (PBS) or with 1.0 SGE and then inoculated with DENV2 at an MOI of 1 for 1 h. (F) Cell migration was observed by light microscopy. (G) Total RNA was extracted, and the expression of DENV2 vRNA and GAPDH was assessed by RT-qPCR 18 hpi. DENV2 vRNA was normalized to GAPDH. (H) Total RNA was extracted from MEFs suspended in collagen matrices, and the expression of DENV2 vRNA was assessed by RT-qPCR 18 hpi and normalized to grams of collagen. Data represent the averages from more than three independent experiments. Student's t tests were performed to analyze statistical significance.