Abstract
Cytomegaloviruses (CMV) have developed various strategies to escape the immune system of the host. One strategy involves the expression of virus-encoded chemokines to modulate the host chemokine network. We have identified in the English isolate of rat CMV (murid herpesvirus 8 [MuHV8]) an open reading frame encoding a protein homologous to the chemokine XCL1, the only known C chemokine. Viral XCL1 (vXCL1), a glycosylated protein of 96 amino acids, can be detected 13 h postinfection in the supernatant of MuHV8-infected rat embryo fibroblasts. vXCL1 exclusively binds to CD4− rat dendritic cells (DC), a subset of DC that express the corresponding chemokine receptor XCR1. Like endogenous rat XCL1, vXCL1 selectively chemoattracts XCR1+ CD4− DC. Since XCR1+ DC in mice and humans have been shown to excel in antigen cross-presentation and thus in the induction of cytotoxic CD8+ T lymphocytes, the virus has apparently hijacked this gene to subvert cytotoxic immune responses. The biology of vXCL1 offers an interesting opportunity to study the role of XCL1 and XCR1+ DC in the cross-presentation of viral antigens.
INTRODUCTION
Chemokines are small chemotactic cytokines that are classified into the CXC, CC, C, and CX3C subfamilies based on the positions of conserved cysteine residues at their N terminus. The C chemokine subfamily is characterized by only one cysteine at the N terminus and contains only one member, XCL1. In humans, two variants (XCL1 and XCL2), which differ by two amino acids, have been reported (1). XCL1, a glycosylated 93-amino-acid mature protein (2), has been shown to be secreted by activated NK cells (3) and CD8+ T cells (4). Murine and human XCL1 chemokines specifically chemoattract a particular subset of dendritic cells (DC) that express XCR1, the only receptor for XCL1 (5, 6). XCR1+ DC excel in antigen cross-presentation, a process in which extracellular antigens are presented by major histocompatibility complex class I (MHC-I) molecules to CD8+ T cells (7–9). Antigen cross-presentation is thought to play a major role in the immune defense against viruses that do not directly infect DC (10, 11). The XCL1-XCR1 interaction facilitates the communication between XCR1+ DC and activated CD8+ T cells or NK cells secreting XCL1 during infection and thereby promotes the differentiation of CD8+ T cells into cytotoxic effector cells (reviewed in reference 12).
Cytomegaloviruses (CMV) belong to the Herpesviridae, a family of large double-stranded DNA viruses that infect a broad spectrum of species and cause lifelong infections in their respective hosts (13). In order to survive successfully and establish latency, CMV have developed various strategies to escape different immune defense mechanisms. One strategy involves the expression of virus-encoded chemokines that interfere with the host chemokine network. It has been speculated that these genes were obtained from the host genome during coevolution and that they contribute to viral dissemination and maintenance (14).
So far, CXC and CC chemokines have been described for rodent, primate, and human CMV (HCMV). Three chemokine-like genes have been described in the HCMV genome: UL128, UL146, and UL147 (15, 16). UL146 and UL147 encode the proteins vCXC-1 and vCXC-2, respectively. Only vCXC-1 has been shown to represent a functional chemokine since it binds to the chemokine receptors CXCR1 and CXCR2 and induces migration of neutrophils to the site of infection (17), a process that has been suggested to facilitate viral dissemination. Whereas one study showed that the UL128 gene product pUL128 blocked migration and induced downregulation of chemokine receptors in monocytes (18), another report demonstrated an opposite effect for pUL128, as it induced migration of peripheral blood mononuclear cells (19), a discrepancy that might be due to N-terminal modifications of the chemokines resulting in different chemotactic behavior. It has also been reported that murid herpesvirus 2 (MuHV2) r129 induces migration of immune cells (20). UL146 and UL147 are restricted to genomes of primate CMVs and have no sequence counterparts in rodent CMV.
CC chemokines have been characterized in rodent CMV, e.g., Maastricht rat CMV (RCMV; MuHV2)-encoded rck-3 (21) and rck-2 (22), murine CMV (MCMV; MuHV1)-encoded chemokine mck-2 (23), and English RCMV (MuHV8)-encoded eck-2 (24). Among these chemokines, MCK-2 is the most extensively studied viral chemokine. MCK-2 has been shown to enhance recruitment of inflammatory monocytes to the site of infection (25). Attracted monocytes inhibit CD8+ T cell activation and cytotoxicity, which results in slower viral clearance (26).
Our analysis of the MuHV8 genome (27) revealed the presence of a C chemokine, located at nucleotide positions 186261 to 186608 toward the right terminus, which to our knowledge is the first viral C chemokine to be reported. The gene product, designated vXCL1, shares extensive homology with the C chemokine XCL1 of rat, mouse, and human. Here, we show that vXCL1 carries a cleavable N-terminal signal sequence that allows secretion from infected cells. Furthermore, vXCL1 functionally resembles host XCL1 since it binds to XCR1+CD4− DC and selectively chemoattracts this particular cell subset. Since murine and human XCR1-expressing DC excel in antigen cross-presentation, vXCL1 might attract this rat DC subset in order to manipulate and disable this important branch of the immune defense.
MATERIALS AND METHODS
Viruses and cell culture.
MuHV8 was propagated on rat embryo fibroblasts (REF) maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 2% fetal bovine serum, 2 mM l-glutamine, and 100 μg/ml penicillin/streptomycin. To determine vXCL1 mRNA and protein expression kinetics, REF were treated 2 h prior to infection or 3 h prior to harvest with 100 μg/ml cycloheximide (Sigma-Aldrich, Taufkirchen, Germany) or 5 μg/ml brefeldin A (Sigma-Aldrich), respectively. In order to generate a MuHV8 mutant lacking vxcl1, a shuttle vector was generated, containing 1.5 kbp of upstream and downstream sequences of vxcl1 using e155 forward (5′-TAGCCGAGACTTCGCACTTC-3′) and reverse (5′-GCCGGAGAGGTGTTTGATTC-3′) primers, and e159 forward (5′-TTCACTACCGAGTGGAACTG-3′) and reverse (5′-GTTCGCAACGAGACCGTCAG-3′) primers, respectively. The vxcl1 open reading frame (ORF) in this shuttle vector was exchanged by a green fluorescent protein (GFP) expression cassette flanked by two LoxP sites. To knock out the vxcl1 ORF within the MuHV8 genome, REF were cotransfected with 2 μg of linearized shuttle vector and 2 μg MuHV8 virion DNA using Polyfect according to the manufacturer's recommendations. The vxcl1 knockout virus was identified by GFP fluorescence and isolated by limiting dilution.
RNA isolation, cDNA synthesis, PCR, and 3′ and 5′ rapid amplification of cDNA ends (RACE).
Isolation of viral and cellular RNA was carried out with the RNeasy Mini Isolation kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). Two million cells were used per RNA isolation column, and remaining DNA contaminations were removed by a 30-min digestion with 20 units of Turbo-DNase (Ambion, Darmstadt, Germany) on the column. For cDNA generation, 1 μg of RNA was incubated for 1 h at 45°C with the following components: 200 U RevertAid H minus reverse transcriptase, 5 μM oligo(dT)18 primer, 1× reaction buffer, 1 mM deoxynucleoside triphosphate (dNTP), and 20 U RiboLock RNase inhibitor (Fermentas, St. Leon-Rot, Germany). To exclude DNA contaminations present in the RNA preparation, cDNA synthesis was additionally carried out with the same components except reverse transcriptase. The reaction was terminated by heating the mixture for 10 min at 70°C. A 1-μl volume of the reaction mixture was applied in a PCR using the Platinum Taq DNA polymerase (Invitrogen, Karlsruhe, Germany) to amplify cDNA with gene-specific primers according to the manufacturer's recommendations. The primer pair vXCL1 universal fwd (5′-ATGCGAGCGGTAATCTTTG-3′) and vXCL1 universal rev (5′-CAGGAACCTGCGTGGGAATA-3′) was used for the amplification of vXCL1 mRNA, the primer pair c-myc fwd (5′-GCCAGAGGAGGAACGAGCT-3′) and c-myc rev (5′-GGGCCTTTTCATTGTTTTCCA-3′) for the amplification of c-myc mRNA, and the primers GAPDH fwd (5′-GGTCGGTGTGAACGGATTTG-3′) and GAPDH rev (5′-GTGAGCCCCAGCCTTCTCCAT-3′) for the amplification of glycerol-3-phosphate dehydrogenase (GAPDH) mRNA.
The 3′-untranslated region (UTR) and the 5′-UTR of vXCL1 were determined with the FirstChoice RLM-RACE kit (Ambion) according to the manufacturer's instructions. vxcl1 gene-specific primers 3′-UTR vXCL1 (5′-CACGAAACCATCTGCGTAAG-3′) and 5′-UTR vXCL1 (5′-AGGAACCTGCGTGGGAATAACTG-3′) were used for amplification of vXCL1-specific mRNA.
Generation of vXCL1-specific MAb vXCL1.11.
For overexpression, the complete coding region of vXCL1 was cloned into the BamHI site of vector pQE-30 (Qiagen) and transformed into Escherichia coli host strain M15 [pREP4] (Qiagen). BALB/c mice were immunized subcutaneously with recombinant protein (50 μg) in complete Freund's adjuvant. Two weeks later, mice were boosted with the same protein in incomplete Freund′s adjuvant by injecting two-thirds of the volume subcutaneously and one-third of the volume intraperitoneally (i.p.). After 2 weeks, sera of immunized mice were screened for antibody titer against the immunogen by enzyme-linked immunosorbent assay (ELISA). The best responders were additionally boosted i.p. with the immunogen dissolved in phosphate-buffered saline (PBS). Three days later, spleen cells were collected and fused with SP2/0 myeloma cells at a ratio of 1:1 after lysis of red blood cells. Cells were seeded on 96-well tissue culture plates in 20% RPMI 1640 medium containing hypoxanthine, aminopterin, and thymidine for hybridoma selection. Cultures were screened for monoclonal antibodies (MAbs) reactive against immunogens by ELISA. Positive mother wells were expanded and cloned. Mice used for immunization were bred and maintained under specific-pathogen-free (SPF) conditions at the Laboratory Mouse Breeding and Engineering Centre (LAMRI) at the Faculty of Medicine, University of Rijeka. All experimental procedures were approved by the Ethics Committee of the University of Rijeka.
Other antibodies, flow cytometry, and recombinant chemokines.
Antibodies recognizing CD45RA (OX-33) and MHCII (OX-6) were from BD Pharmingen, anti-CD4 (W3/25) antibodies from AbDSerotec, and anti-CD103 (OX62) antibodies from Biolegend. Mouse XCR1-specific monoclonal antibody (MARX10 [9], cross-reacting with rat XCR1) and goat serum directed against murine XCL1 (cross-reacting with vXCL1) were generated in the laboratory of R.A.K. Polyclonal rabbit anti-goat immunoglobulin-horseradish peroxidase (HRP) and goat anti-mouse immunoglobulin-HRP were obtained from Dako. Flow cytometry data were acquired on an LSR II or FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA) using Facsdiva or Cell Quest Pro software (BD Biosciences), respectively. Final examination and compensation of the data were carried out using FlowJo software (Tree Star, Ashland, OR, USA). vXCL1 and rat XCL1 containing an affinity tag at their respective carboxy termini were cloned into the expression vector pRmHa-3 (28) and expressed in stable Drosophila SL-3 cell transfectants (L. Voss, H.W. Mages, and R.A. Kroczek, unpublished data).
Sandwich ELISA.
Maxisorp 96-well microtiter plates (Nalgene Nunc, Roskilde, Denmark) were coated with 100 μl MAb vXCL1.11 (final concentration, 1 μg/ml) diluted in coating buffer (0.1 M NaHCO3, Na2CO3, pH 9.5) overnight at 4°C. Wells were washed five times with washing buffer (1× PBS containing 0.1% [vol/vol] Tween 20) and blocked with blocking buffer (1% bovine serum albumin [BSA], wt/vol, in washing buffer) for 2 h at room temperature. After five washes with washing buffer, 100 μl of test sample and serial dilutions of recombinant vXCL1 were applied per well and incubated for 2 h at room temperature. Unbound proteins were removed by rinsing five times with washing buffer, followed by 1 h of incubation with cross-reactive goat serum directed against murine XCL1 (1:500 dilution) in blocking buffer at room temperature. Polyclonal anti-goat immunoglobulin-HRP was diluted in blocking buffer (final concentration, 0.2 μg/ml) and added after rinsing five times with washing buffer for 1 h at room temperature. After 10 additional washes, 50 μl of TMB Plus (Kem En Tec, Taastrup, Denmark) was added to each well and incubated for 5 min in the dark. The reaction was stopped by adding 50 μl of 0.5 N H2SO4, and absorbance was determined at 450 nm using a Spectrafluor Plus (Tecan, San Jose, CA, USA) microplate reader.
SDS-PAGE, immunoblotting, and posttranslational modification analysis.
To analyze N-glycosylation of vXCL1, lysates of MuHV8-infected REF (20 μg of total protein of REF infected at a multiplicity of infection [MOI] of 10) were denatured for 10 min in 1× glycoprotein denaturing buffer (NEB, Ipswich, MA, USA) and then digested with 1 U PNGaseF (NEB) for 1.5 h at 37°C using 1× G7 reaction buffer (NEB) and 1% Nonidet P-40. The digested protein lysate was then mixed with 4× SDS loading buffer (0.3 M Tris, 12% SDS, 40% [vol/vol] glycerol, 0.3 M dithiothreitol [DTT], 0.02 bromophenol blue) and separated electrophoretically on a 15% Tris-Tricine SDS gel using the prestained PageRuler ladder (Fermentas). Afterwards, the gel was blotted onto a Hybond ECL nitrocellulose membrane (GE Healthcare, Munich, Germany) with 0.8 mA per cm2 and stained with MAb vXCL1.11 using the ECL Western blotting reagents (GE Healthcare).
Mass spectrometric peptide analysis.
vXCL1 produced in insect cells was purified by heparin Sepharose affinity chromatography. Peptides from the gel-purified protein were obtained by trypsin and endoproteinase Glu-C in-gel digestion as described previously (29), and peptide masses were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF/MS) using an Ultraflex-II TOF/TOF instrument (Bruker Daltonics, Bremen, Germany) equipped with a 200-Hz solid-state Smart beam laser. The mass spectrometer was operated in the positive reflector mode; mass spectra were acquired over an m/z range from 600 to 4,000. CHCA (α-cyano-4-hydroxycinnamic acid) was used as the matrix, and samples were spotted using the dried-droplet technique. Tandem MS (MS/MS) spectra of selected peptides were acquired in LIFT mode (30).
Cell isolation.
Spleens of Lewis rats were cut into small pieces and digested with 500 μg/ml collagenase D (Roche, Penzberg, Germany) and 20 μg/ml DNase I (Roche) in RPMI 1640 containing 2% fetal calf serum (FCS; Pan Biotech GmbH, Regensburg, Germany) for 25 min at 37°C and shaking at 200 rpm. After addition of 10 mM EDTA and incubation for 5 min at 37°C, cells were filtered through a 100-μm nylon sieve (BD Pharmingen) followed by NycoPrep density gradient (1.073 g/ml) for 10 min at 1,700 × g and 4°C. The DC-enriched interphase was recovered and used for chemokine binding assays. For chemotaxis experiments, rat DC were enriched by magnetic cell sorting using CD103 microbeads according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany).
Chemotaxis assay.
Migration of CD103-enriched splenocytes was analyzed in a migration assay as described elsewhere (31). In brief, 1 × 105 to 5 × 105 CD103-enriched rat splenocytes were resuspended in chemotaxis medium (RPMI 1640, 1% BSA, 50 μM β-mercaptoethanol, 100 μg/ml penicillin-streptomycin) and placed in the upper chamber of a 24-well 6.5-mm transwell system (Corning, Salt Lake City, UT, USA). The lower chamber was filled with chemotaxis medium containing the chemokine of interest, no chemokine as a control, or supernatant of infected REF. Cells were incubated for 2.5 h at 37°C and 5% CO2. To examine the effect of pertussis toxin (PTX) on cell migration, CD103-enriched splenocytes were treated with 100 ng/ml PTX (Sigma-Aldrich, Taufkirchen, Germany) for 2 h at 37°C and 5% CO2 and then washed three times with chemotaxis medium prior to migration analysis. Cells migrating into the lower chamber were analyzed by flow cytometry. T cells (CD3+, MHCII−), B cells (CD45RA+, MHCII+), CD4+ DC (MHCII+, CD103+, CD4+), and CD4− DC (MHCII+, CD103+, CD4−) were identified by flow cytometry using the indicated markers. Numbers of migrated cells and input cells were determined by counting cells over a time frame of 300 s in a defined volume. The calculation of the percentage of migrated cells was based on the number of input cells: (number of migrated cells/number of input cells) × 100.
RESULTS
MuHV8 encodes a C chemokine homologue.
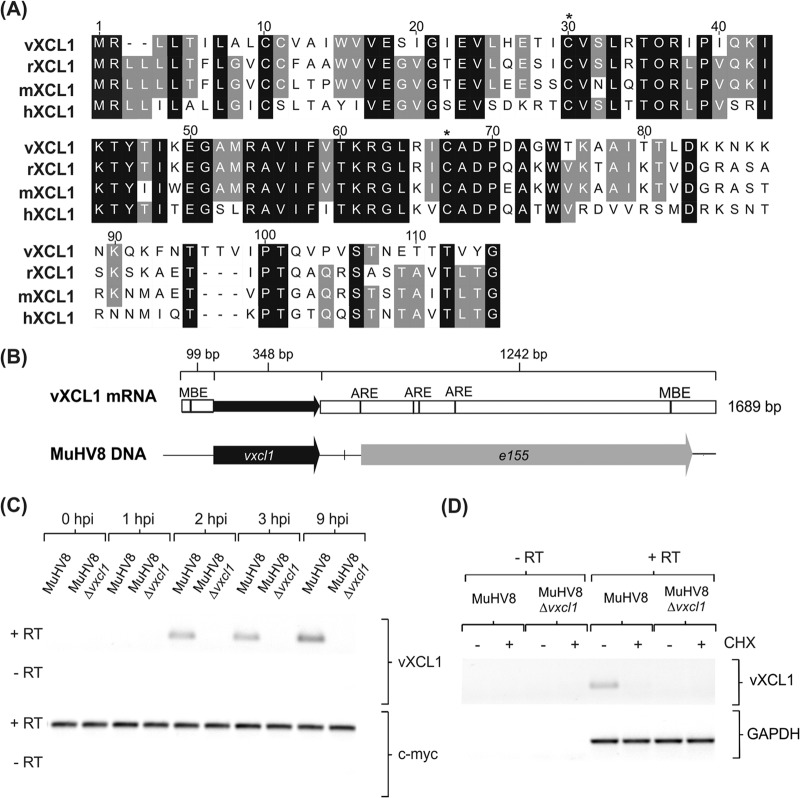
Genome analysis of MuHV8 identified a 348-bp ORF between viral genes e155 and e159 encoding a putative 115-amino-acid protein. BLAST analysis at the protein level revealed 64% identity (73% homology) with the C chemokine XCL1 of Rattus norvegicus, 58% identity with mouse XCL1, and 46% identity with human XCL1 (Fig. 1A). Given the high degree of identity and homology to XCL1 and the conserved cysteines at positions 30 and 67, characteristic of C chemokines, this putative protein was designated viral XCL1 (vXCL1). The highest divergence between vXCL1 and XCL1 was observed at the C terminus, whereas the N terminus and the core of the protein were conserved.
FIG 1.
Characterization of MuHV8-encoded C chemokine vXCL1. (A) Muscle alignment (Geneious version 5.5.7) of amino acid sequences of vXCL1 and rat, mouse, and human XCL1. Identical amino acids are indicated by a black background and similar amino acids by a gray background, and lack of similarity is indicated by a white background. Cysteine positions characteristic of C chemokines are indicated by asterisks. Numbers refer to the amino acid positions of the viral chemokine. Accession numbers for the sequences depicted are JX867617.1 (vXCL1, MuHV8), NP_599188 (rXCL1, Rattus norvegicus XCL1), NP_032536 (mXCL1, Mus musculus XCL1), and NP_002986 (hXCL1, Homo sapiens XCL1). (B) Analysis of vXCL1 mRNA. Total RNA from MuHV8-infected REF was isolated 24 h postinfection (hpi) and 3′- and 5′-UTRs were determined. Musashi binding elements (MBE) were detected using UTRScan, and AU-rich elemenst (ARE) were identified by scanning for an AUUUA element with CloneManager (Sci-Ed software, Cary, NC, USA). (C) Time course of MuHV8 vXCL1 expression. Total RNA from wild-type MuHV8 and MuHV8 Δvxcl1-infected cells was harvested at indicated time points, reverse transcribed with oligo(dT) primers, and amplified with vxcl1-specific primers. (D) Cycloheximide (CHX) was used to discriminate between immediate early and early gene expression. MuHV8 Δvxcl1 was included as a negative control. The experiment was carried out at least twice with similar results. c-myc and GAPDH served as controls. RT, reverse transcriptase.
Rat, murine, and human xcl1 each contain three exons that are transcribed into a single mRNA (1, 32). In contrast, vXCL1 mRNA is unspliced. To investigate vXCL1 expression, mRNA of MuHV8-infected rat embryo fibroblasts (REF) was harvested and the 3′-UTR and 5′-UTR were determined by rapid amplification of cDNA ends (RACE). MuHV8 vxcl1 encodes a transcript of 1,689 bp consisting of a 99-bp 5′-UTR, a 348-bp ORF, and a 1,242-bp 3′-UTR (Fig. 1B). Computational analysis detected four AU-rich elements (ARE), one Musashi binding element (MBE) in the 3′-UTR, and one MBE motif in the 5′-UTR of vXCL1 mRNA.
If vXCL1 were to be involved in immune evasion, precise timing of viral gene expression to bypass certain host immune responses would be expected; we therefore analyzed vxcl1 expression kinetics. Semiquantitative reverse transcription (RT)-PCR revealed that vXCL1 mRNA is already expressed 2 h postinfection and follows early kinetics (Fig. 1C). To clarify if vXCL1 mRNA is produced immediate early or early, REF were treated with cycloheximide prior to infection. vXCL1 mRNA was not detected upon treatment with the protein synthesis inhibitor (Fig. 1D), indicating that vxcl1 is expressed early after infection in a protein-dependent fashion.
vXCL1 is a posttranslationally modified, secreted protein.
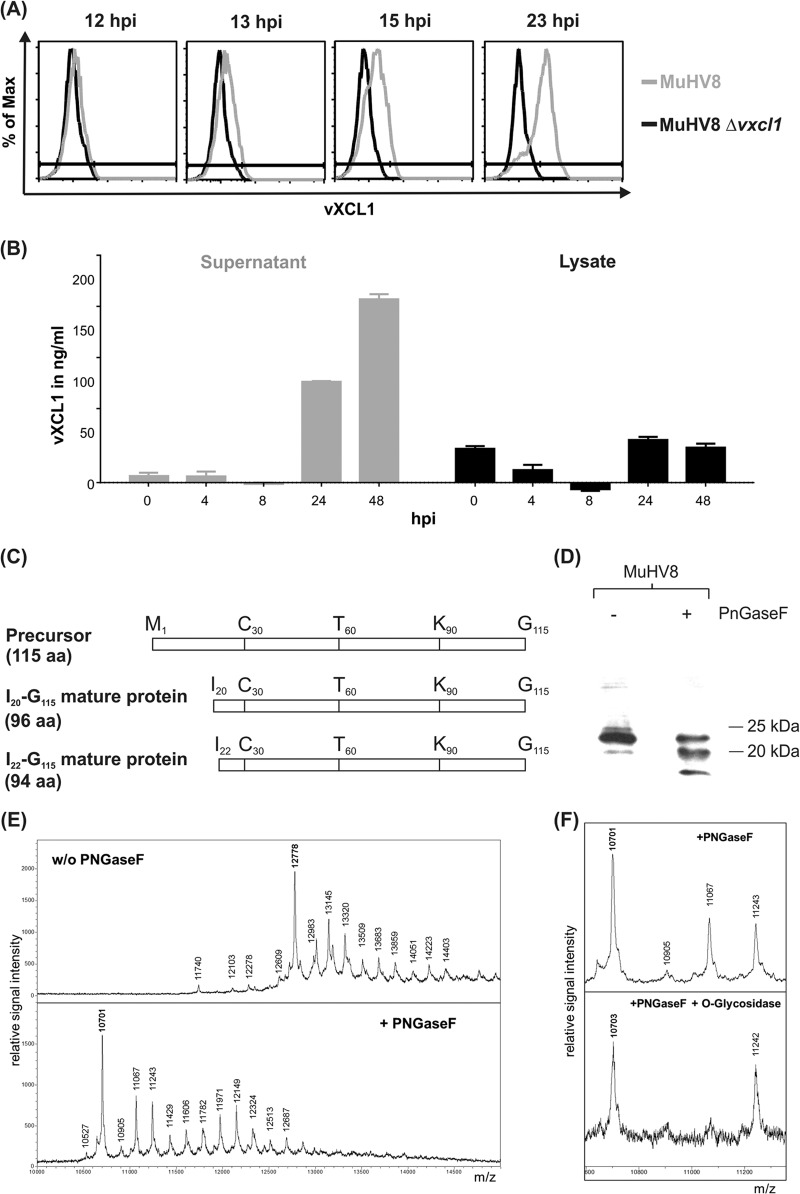
To examine if and at what time postinfection vXCL1 is secreted, flow cytometry and sandwich ELISAs were carried out using a monoclonal antibody directed against vXCL1 (MAb vXCL1.11). As shown in Fig. 2A, vXCL1 could be detected in infected cells by flow cytometry at 13 h postinfection and accumulated thereafter in the presence of brefeldin A. A quantitative comparison of vXCL1 protein in lysates and supernatants of infected cells by ELISA revealed that vXCL1 is enriched over time in the supernatant of infected REF (Fig. 2B). Only small amounts of viral protein were detected throughout the course of infection in cell lysate.
FIG 2.
Analysis of vXCL1 protein expression and posttranslational modification. (A) Protein expression kinetics of vXCL1. MuHV8-infected REF were treated 3 h before harvest with brefeldin A, harvested at indicated time points, and analyzed by flow cytometry using MAb vXCL1.11, which specifically recognizes vXCL1 (gray line). REF infected with MuHV8 Δvxcl1 served as a control (black line). (B) Secretion of vXCL1. Supernatants (gray bars) and lysates (black bars) of infected REF were collected at indicated time points and tested for the presence of vXCL1 by sandwich ELISA. Values were normalized by subtracting the mean of the value obtained with MuHV8 Δvxcl1-infected cells. Results shown are from two independent experiments. Error bars represent means ± standard deviations (SD). (C) Schematic overview of the precursor protein and the two variants of mature vXCL1 determined by mass spectrometric analysis. aa, amino acids. (D) N-glycosylation variants of vXCL1. Lysates of MuHV8-infected REF were digested for 1.5 h in the presence or absence of PNGaseF and separated electrophoretically on a 15% Tris-Tricine polyacrylamide gel, followed by blotting and detection using MAb vXCL1.11. (E) Mass spectrometric analysis of vXCL1 N-glycosylation. PNGase F treatment resulted in a main peak shift from 12,778 to 10,701, the expected mass of the vXCL1 protein. (F) Mass spectrometric analysis of vXCL1 O-glycosylation. The mass peak at 11,067 in the N-deglycosylated sample (top panel) could be eliminated by O-glycosidase (bottom panel).
Chemokines are usually generated as precursor proteins containing an N-terminal signal peptide of approximately 20 residues (e.g., in human XCL1 [2]). Cleavage of the signal peptide yields the mature protein, which is secreted. In order to characterize the N terminus of MuHV8 vXCL1 experimentally, the viral protein was expressed in insect cells and harvested from the supernatant by heparin affinity chromatography. Following purification, recombinant vXCL1 was analyzed by peptide mass fingerprinting. MALDI mass spectrometric analysis of secreted vXCL1 detected isoleucine20 (I20) as the N-terminal amino acid, indicating that the precursor protein is cleaved between serine19 (S19) and isoleucine20 (I20), resulting in a 96-residue mature protein. Additionally, the analysis revealed the presence of another mature protein starting with isoleucine22, although this form was less abundant. Figure 2C summarizes the structures of the two mature protein variants.
Chemokine glycosylation has been implicated in chemokine folding and stability, thereby influencing the interaction with the respective receptor (33). Glycosylation was found to modify human XCL1 in such a way that different molecular sizes become apparent in Western blot analysis (2). To clarify whether different posttranslationally modified versions of vXCL1 exist, lysates of REF infected with MuHV8 were analyzed by Western blotting after digestion with PNGaseF, an amidase cleaving N-linked oligosaccharides from asparagines. As shown in Fig. 2D, PNGaseF treatment led to a size shift of vXCL1, indicating the presence of N-linked sugars attached to the viral protein. These data were corroborated by mass spectrometric analyses. PNGase F treatment resulted in a shift of the main peak from 12,778 to 10,701, indicating that vXCL1 is N-glycosylated (Fig. 2E). The observed mass shift of 2,077 could best be explained by attachment of a Man3GlcNac2Fuc moiety (m = 1,039) to each of the two potential N-glycosylation sites at N94 and N108. After removal of N-linked oligosaccharides, the mass heterogeneity persisted, presumably due to heterogeneous O-glycosylation. In the spectrum of the N-deglycosylated sample, the mass peak at 11,067 pointed to an additional HexNAc-Hex, compatible with a mucin-type O-glycan core structure. This peak could be eliminated by O-glycosidase, also known as endo-α-N-acetylgalactosaminidase, an enzyme that preferentially catalyzes the removal of O-linked disaccharides from glycoproteins, while it does not attack more-complex O-glycan structures. These findings suggest that vXCL1 is additionally O-glycosylated (Fig. 2F).
vXCL1 is a chemoattractant for CD4− DC, but not for CD4+ DC, T cells, or B cells.
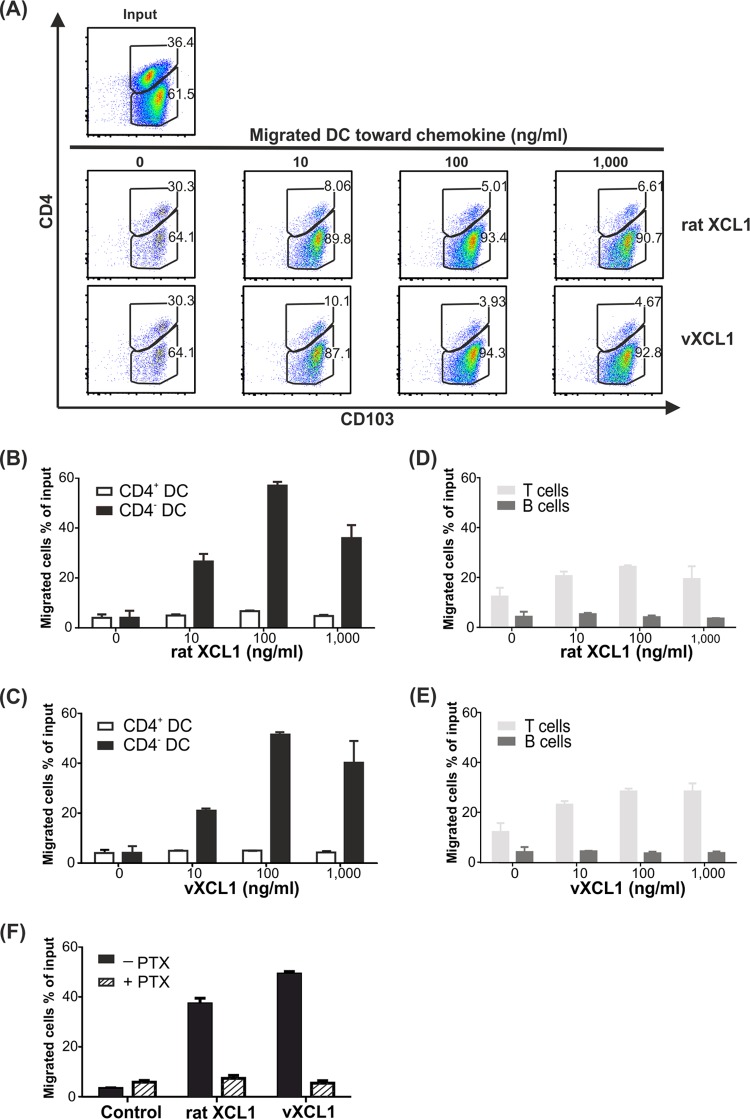
Murine and human XCL1 chemokines exert their biological function by attracting XCR1-expressing DC. We therefore tested vXCL1 for chemotactic capacity on rat DC, which were defined as CD103+MHCII+ (34), and used rat XCL1 as a positive control. DC were obtained by collagenase digestion of splenic tissue followed by NycoPrep density gradient centrifugation and magnetic sorting of CD103+ cells. The enriched (60%) DC population (composed of 36% CD4+ and 62% CD4− DC; Fig. 3A, input) was placed in the upper chamber of a transwell system. Only very low background migration was observed with CD4− DC and CD4+ DC fractions (less than 5% of input cells) in the absence of rat XCL1 or vXCL1. With increasing concentrations of rat XCL1 and vXCL1, selective migration of CD4− DC occurred while CD4+ DC essentially failed to migrate (Fig. 3B and C). A concentration of 100 ng/ml of either vXCL1 or rat XCL1 yielded the strongest migration, approximately 60% of CD4− DC. vXCL1 and rat XCL1 did not induce migration of T or B cells (Fig. 3D and E). These results demonstrate that vXCL1 is a bona fide chemokine selectively attracting CD4− DC but not CD4+ DC. Since chemokine receptors have been shown to signal via PTX-sensitive Gαi proteins (35), we investigated if the latter play a role in rat XCR1 signaling. DC were preincubated with 100 ng/ml PTX and analyzed for their ability to migrate toward vXCL1 or rat XCL1 (Fig. 3F). Migration of CD4− DC toward vXCL1 and rat XCL1 was blocked by PTX, indicating that XCR1 signaling involves Gαi protein.
FIG 3.
Analysis of vXCL1 chemotactic activity. (A) Magnetically sorted splenic DC were tested for migration toward different concentrations of vXCL1 and rat XCL1 (10 to 1,000 ng/ml) in a transwell assay. DC subpopulations were defined based on expression of CD103 and CD4. The dot plots represent the numbers of CD4− DC and CD4+ DC that have migrated into the lower chamber. (B and C) Proportions of migrated CD4− and CD4+ DC in response to different concentrations of rat XCL1 and vXCL1. (D and E) Proportions of migrated T and B cells toward rat XCL1 and vXCL1. (F) Pertussis toxin (PTX) blocks migration of CD4− DC toward rat XCL1 and vXCL1. No migration is detected in the absence of chemokine (control). All experiments were performed twice. Error bars represent means ± standard errors of the means (SEM).
MuHV8-infected cells secreting vXCL1 attract CD4− DC.
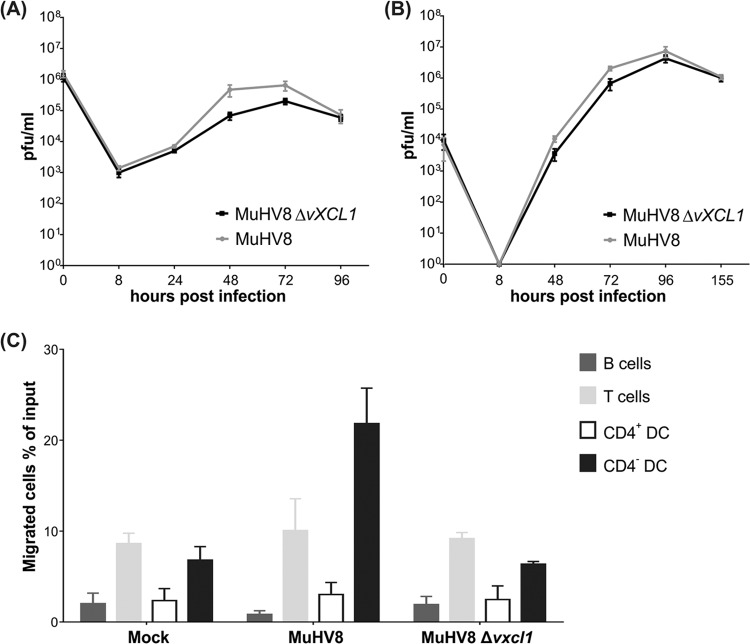
Since recombinant vXCL1 induced migration of DC, it was expected that supernatant harvested from cells infected with wild-type virus but not virus with deletion of vxcl1 (Δvxcl1) would also result in DC migration. However, to rule out that differences in replication kinetics between wild-type and Δvxcl1 MuHV-8 cause variations in DC migration, the growth behaviors of the viruses were analyzed and found to be similar (Fig. 4A and B).
FIG 4.
vXCL1 is dispensable for viral growth in cell culture and attracts CD4− DC. Growth of wild-type MuHV8 and MuHV8 Δvxcl1: REF were infected at MOIs of 5 (A) and 0.05 (B), respectively, and titers were determined at the indicated time points by plaque assay. Results were obtained from three independent experiments. (C) The ability of vXCL1 to induce chemotaxis of different rat cell subsets was analyzed with supernatants of mock-infected, wild-type MuHV8-infected, and MuHV8 Δvxcl1-infected REF. Results were obtained from two independent experiments. Error bars represent means ± SD.
Forty-eight hours postinfection, supernatants of mock-infected, MuHV8 wild-type-infected and Δvxcl1 mutant-infected REF were analyzed in chemotaxis assays (Fig. 4C). Supernatants of cells infected with Δvxcl1 MuHV8 failed to induce migration of CD4− DC above the background compared with supernatants obtained from mock-infected REF. In contrast, supernatants of MuHV8 wild-type-infected REF induced chemotaxis in more than 20% of CD4− DC. None of the supernatants induced migration of T or B cells. Taken together, these data indicate that vXCL1 generated during infection selectively attracts CD4− DC.
Both vXCL1 and rat XCL1 bind to XCR1+CD4− DC.
In the mouse and humans, expression of XCR1 is restricted to a subset of DC (9, 31, 36). Since XCR1 is the only receptor for murine and human XCL1, we tested whether vXCL1 and rat XCL1 interact with rat XCR1. For these experiments, we used MAb MARX10, which specifically recognizes murine XCR1 (9) in a nonblocking fashion and cross-reacts with rat XCR1 (E. Hartung, H. Geyer, S. Voigt, and R. A. Kroczek, unpublished data). CD4− and CD4+ splenic DC were stained with MARX10 and costained with either rat XCL1 or vXCL1 (both tagged with a fluorochrome). As shown in Fig. 5, vXCL1 bound exclusively to CD4− DC expressing XCR1 (bold square). Neither CD4+ DC nor T or B cells exhibited any binding of the viral chemokine. Rat XCL1 was tested in parallel and demonstrated a very similar staining pattern.
FIG 5.
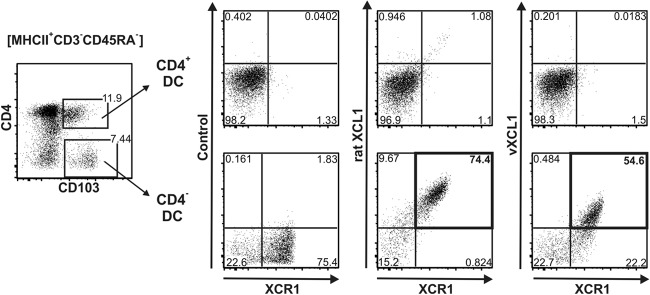
Analysis of rat XCL1 and vXCL1 binding to XCR1. Splenic rat DC (MHCII+CD103+CD3−CD45RA−) were gated into CD4+ (upper panels) and CD4− (lower panels) DC subsets and analyzed for binding of XCL1, vXCL1, and MAb MARX10. Both rat XCL1 and vXCL1 selectively bound to DC expressing XCR1 (bold frames); the inset numbers give the proportion of double-positive cells. One representative experiment of three is shown.
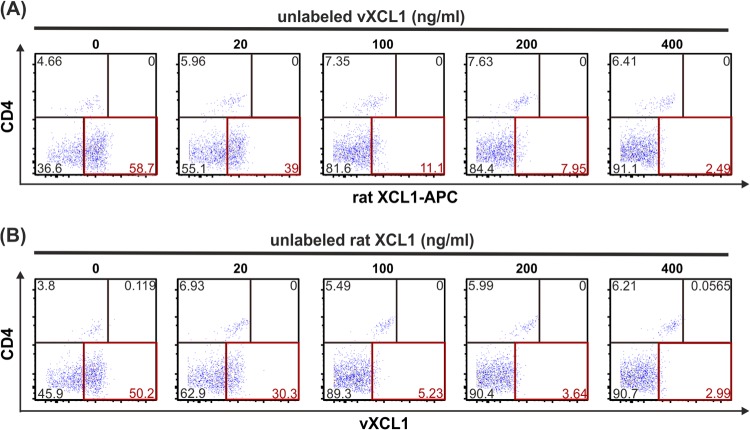
Since rat XCL1 and vXCL1 are highly similar in their amino acid sequence and induced chemotaxis and are bound to the same cell subset, we tested whether they share the same receptor, XCR1. To address this question, enriched DC were incubated with rat XCL1-APC in the presence of increasing concentrations of unlabeled vXCL1 and vice versa. Figure 6A shows that binding of rat XCL1-APC to CD4− DC was effectively competed by unlabeled vXCL1. Analogous results were obtained when DC were incubated with vXCL1-APC in the presence of unlabeled rat XCL1 (Fig. 6B). These findings indicate that both chemokines bind to the same receptor on CD4− DC.
FIG 6.
Competitive binding of vXCL1 and rat XCL1 to rat CD4− DC. NycoPrep gradient-enriched rat DC were incubated with 400 ng/ml rat XCL1-APC in the presence of different concentrations of unlabeled vXCL1 (A) or with 400 ng/ml vXCL1-APC in the presence of different concentrations of unlabeled rat XCL1 (B), washed, and analyzed for binding of the respective fluorophore-tagged chemokine to rat DC (CD103+MHCII+). In both instances, binding of the labeled chemokine was effectively competed by the unlabeled chemokine on CD4− DC (lower right quadrants). One representative experiment of two is shown.
DISCUSSION
CMV encodes several proteins that interfere with the chemokine network of the host. These proteins include molecules that act as chemokines (21), as chemokine-binding proteins (37), or as chemokine receptors (38, 39). In our study, we identified and characterized a C chemokine encoded by MuHV8, vXCL1, to our knowledge the first reported viral C chemokine. We could not identify sequence homologues in any other virus by database screening.
BLAST analysis of the vXCL1 amino acid sequence revealed high similarity to the only known C chemokine expressed in rat, mouse, and humans, XCL1. XCL1 in various species and vXCL1 have a similar length, of 114 to 115 amino acids, in their immature form. Both mouse and rat xcl1 have three coding exons. Rat xcl1 (GenBank Gene ID, 171371) has a size of 3.44 kbp with a transcript length of 345 bp, and mouse xcl1 (GenBank Gene ID, 16963 [40]) has a size of 3.88 kbp with a transcript of 523 bp. The vxcl1 transcript of 1.6 kbp (including a 1.2-kbp 3′-UTR) originates from a continuous viral gene segment and contains one MBE motif and various AU-rich elements. In contrast to vxcl1, the MuHV8-encoded CC chemokine eck-2 was acquired as an unspliced transcript with intron/exon boundaries similar to mck-2 in MCMV (24). Possibly, acquisition of unspliced transcripts enables different levels of regulation of the CC chemokines, whereas this might not occur in the case of vxcl1.
Expression of immunomodulatory CMV genes is precisely timed to compromise cellular defense strategies (41). We therefore carefully analyzed both vXCL1 mRNA and protein expression kinetics. Semiquantitative RT-PCR revealed that vXCL1 mRNA is expressed 2 h postinfection and therefore follows early kinetics. In contrast to these results, intracellular vXCL1 protein could be detected only 13 h after infection. Since the vXCL1 transcript contains various RNA-binding protein motifs, it is conceivable that vXCL1 mRNA translation is inhibited early during infection through the interaction with RNA-binding proteins. Moreover, vXCL1 mRNA contains more AU-rich elements and also other potential binding sites for proteins and microRNAs than host rat XCR1. These sites might influence translation of vXCL1 mRNA and could serve as an explanation for the discrepancy between mRNA production and the onset of protein expression.
Correct cleavage of the signal peptide in the conversion of an immature chemokine into its mature form is critical for receptor binding and function (42). Since vXCL1 was secreted and enriched in the supernatant of infected REF, we performed mass spectrometric analysis to determine the N terminus of the mature protein. The supernatants contained two variants of vXCL1: an abundant mature protein spanning from isoleucine20 to glycine115 and a less abundant truncated protein ranging from isoleucine22 to glycine115. In addition, vXCL1 was shown to contain N-linked sugars since PNGaseF treatment resulted in a size shift as detected by Western blotting and mass spectrometric analyses. The vXCL1 amino acid sequences possess two N-glycosylation motifs at which N-glycosylation could take place: Asn94-Thr95-Thr96 and Asn108-Glu109-Thr110. The N-linked-sugars of vXCL1 might improve interaction with the chemokine receptor, increase the stability and thus the biological activity of the chemokine, or help to mask antigenic sites of the viral molecule. Interestingly, human and murine XCL1 presumably carry O-linked sugars at their respective C termini (2, 43). NetOGlyc analysis predicted several potential O-glycosylation sites for vXCL1 at the C terminus, and mass spectrometric analysis confirmed that vXCL1 possesses an O-linked sugar.
Murine and human XCL1 have been shown to act as chemoattractants on cross-presenting DC (9, 31, 36), while there are no data available for rat XCL1. We therefore compared vXCL1 and rat XCL1 in their ability to induce migration of CD4− DC, CD4+ DC, B cells, and T cells. Our data demonstrate that only CD4− DC migrate in the presence of either the host or the viral chemokine. Similar findings were obtained when supernatants of infected REF were used in chemotaxis assays: only supernatants of MuHV8-infected REF but not those of the Δvxcl1mutant-infected cells attracted CD4− DC. A revertant virus was not employed in these assays because complete genome sequencing of the MuHV8 Δvxcl1 knockout virus and comparison with wild-type MuHV8 showed the desired sequence divergence only in the vxcl1 region but not elsewhere in the genome. Rat CD4− DC are thought to be the equivalent of murine CD8+ DC (44, 45) since they both produce large amounts of interleukin 12 (45), efficiently phagocytose apoptotic cells, and are located in the red pulp and T cell area of the spleen (46). The finding that both rat XCL1 and vXCL1 selectively induce migration of only CD4− DC further supports the concept that these DC are the equivalents of human (31, 47) and murine (9) cross-presenting DC.
The G protein-coupled receptor XCR1 was shown to be expressed only on cross-presenting DC in mice (9) and was identified as the only interaction partner of XCL1 in the human and murine systems. Thus, XCR1 is the most prominent candidate as a vXCL1 and rat XCL1 interaction partner. The monoclonal antibody MARX10 directed against mouse XCR1 (9) cross-reacts with rat XCR1 and therefore served as a valuable tool to analyze the correlation between chemokine binding and receptor expression. Approximately 80% of rat CD4− DC expressed XCR1, which correlates with the murine system, where 70 to 85% of spleen-derived CD8+ DC were shown to express XCR1 (9, 36). Flow cytometry studies revealed that vXCL1 and rat XCL1 bound selectively to CD4−, XCR1-expressing DC. Chemokine binding to CD4+ DC was not observed, which is in accordance with our chemotaxis data.
Upon ligand binding, intracellular G proteins are dissociated into GTP-bound subunits, and a signal cascade that results in cell migration is initiated. Since little is known about rat XCR1-mediated signaling, we addressed the question if vXCL1- or rat XCL1-mediated migration can be inhibited by treatment with PTX, an agent that modifies Gαi proteins, and if this would prevent Gαi protein interaction with XCR1 and abolish DC migration. Indeed, preincubation with PTX inhibited CD4− DC migration, suggesting that rat XCR1 is linked with a Gαi protein.
Since both rat XCL1 and vXCL1 bound to XCR1+CD4− DC, it was likely that the two chemokines shared the interaction partner. However, it was not clear if XCR1 is the only interaction partner or if another receptor coexpressed with XCR1 is targeted. To address this question, we examined binding of rat XCL1-APC to rat DC subsets in the presence of different concentrations of unlabeled vXCL1 and vice versa. Competitive binding studies revealed that binding of APC-labeled rat XCL1 was abolished in the presence of unlabeled vXCL1 and vice versa, implying that vXCL1 binds to the same surface molecule, i.e., XCR1.
The data presented here show that vXCL1 (i) has chemokine-like properties, (ii) binds to and attracts CD4− rat DC that express XCR1, and (iii) functionally resembles rat XCL1. At first glance, the attraction of XCR1+ DC seems rather unfavorable for the virus. In mice and humans, XCR1-expressing DC are capable of antigen cross-presentation and thus possess a key role in controlling viruses (10, 11). In the mouse model, CD8+ cross-presenting DC could be infected with MCMV at a low percentage (48, 49). Infection of DC was shown to be accompanied by reduced surface expression of MHC-I and -II molecules (50–52), altered cytokine and chemokine receptor expression (53), and downregulation of molecules required for T and NK cell proliferation (48, 50, 52). Since DC are important for regulating and controlling T and NK cell responses (54, 55) and the latter play a major role in controlling CMV infection (56–60), MuHV8 might infect and functionally paralyze this DC subset by vXCL1 attraction in order to impair antiviral responses. Further, immature DC have been shown to be an important reservoir of latent CMV from which reactivation can occur (61, 62), but it remains to be determined if vXCL1 has a role in the establishment of a latent infection.
Alternatively, it is possible that cross-presenting DC regulate T cell activation and cytotoxicity without being infected and that vXCL1 functions in a manner analogous to that of the MCMV-encoded CC chemokine MCK-2. MCK-2 attracts inflammatory monocytes to the site of infection, where they compromise virus-specific CD8+ T cells and thus contribute to viral persistence (26). Possibly, vXCL1 attracts XCR1+ DC to the site of infection and locally disturbs cooperation with CD8+ T cells, thereby compromising the adaptive immune response. However, it remains to be shown if the attracted XCR1+ DC subset is able to cross-present antigen. Eventually, the influence of MuHV8 on this potentially cross-presenting DC subset and possible vXCL1 interference with the host immune system will have to be evaluated in in vivo studies.
So far, all CMV-encoded chemokines have been classified as either CXC or CC chemokines and have been shown to attract neutrophils (17), monocytes (25), macrophages (63), and CD4+ T cells (20). vXCL1 is the first reported CMV-encoded C chemokine that targets XCR1+CD4− DC. This might imply an important physiological function, since Kaposi's sarcoma-associated herpesvirus (KSHV) has also been shown to encode two chemokines, vCCL2 and vCCL3, which likewise target XCR1 at different time points during infection (64, 65). The observation that both MuHV8 and KSHV exploit XCR1 with different virally encoded chemokines suggests a substantial importance of this receptor in the defense against herpesviruses.
Since MuHV8 is so far the only known virus to encode an XCL1 homologue, the MuHV8-rat model appears to be particularly useful in analyzing the biological function of vXCL1 and its interference with the endogenous XCL1 molecule. Thus, this model offers a unique opportunity to study the involvement of XCL1 in antiviral defenses and might be promising to provide clues for novel antiviral strategies.
ACKNOWLEDGMENTS
We thank Marina Babić as well as Brigitte Dorner and Martin Dorner for expert help.
S.V. is supported by the Deutsche Forschungsgemeinschaft (VO 774/7).
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Yoshida T, Imai T, Takagi S, Nishimura M, Ishikawa I, Yaoi T, Yoshie O. 1996. Structure and expression of two highly related genes encoding SCM-1/human lymphotactin. FEBS Lett. 395:82–88. 10.1016/0014-5793(96)01004-6 [DOI] [PubMed] [Google Scholar]
- 2.Dorner B, Müller S, Entschladen F, Schröder JM, Franke P, Kraft R, Friedl P, Clark-Lewis I, Kroczek RA. 1997. Purification, structural analysis, and function of natural ATAC, a cytokine secreted by CD8(+) T cells. J. Biol. Chem. 272:8817–8823. 10.1074/jbc.272.13.8817 [DOI] [PubMed] [Google Scholar]
- 3.Dorner BG, Smith HR, French AR, Kim S, Poursine-Laurent J, Beckman DL, Pingel JT, Kroczek RA, Yokoyama WM. 2004. Coordinate expression of cytokines and chemokines by NK cells during murine cytomegalovirus infection. J. Immunol. 172:3119–3131 http://www.jimmunol.org/content/172/5/3119 [DOI] [PubMed] [Google Scholar]
- 4.Dorner BG, Scheffold A, Rolph MS, Hüser MB, Kaufmann SH, Radbruch A, Flesch IE, Kroczek RA. 2002. MIP-1alpha, MIP-1beta, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc. Natl. Acad. Sci. U. S. A. 99:6181–6186. 10.1073/pnas.092141999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heiber M, Docherty JM, Shah G, Nguyen T, Cheng R, Heng HH, Marchese A, Tsui LC, Shi X, George SR, O'Dowd BF. 1995. Isolation of three novel human genes encoding G protein-coupled receptors. DNA Cell. Biol. 14:25–35. 10.1089/dna.1995.14.25 [DOI] [PubMed] [Google Scholar]
- 6.Yoshida T, Imai T, Kakizaki M, Nishimura M, Takagi S, Yoshie O. 1998. Identification of single C motif-1/lymphotactin receptor XCR1. J. Biol. Chem. 273:16551–16554. 10.1074/jbc.273.26.16551 [DOI] [PubMed] [Google Scholar]
- 7.Pooley JL, Heath WR, Shortman K. 2001. Cutting edge: intravenous soluble antigen is presented to CD4 T cells by CD8− dendritic cells, but cross-presented to CD8 T cells by CD8+ dendritic cells. J. Immunol. 166:5327–5330 http://www.jimmunol.org/content/166/9/5327.long [DOI] [PubMed] [Google Scholar]
- 8.den Haan JM, Lehar SM, Bevan MJ. 2000. CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192:1685–1696. 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachem A, Hartung E, Güttler S, Mora A, Zhou X, Hegemann A, Plantinga M, Mazzini E, Stoitzner P, Gurka S, Henn V, Mages HW, Kroczek RA. 2012. Expression of XCR1 characterizes the Batf3-dependent lineage of dendritic cells capable of antigen cross-presentation. Front. Immunol. 3:214. 10.3389/fimmu.2012.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rock KL, Shen L. 2005. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunol. Rev. 207:166–183. 10.1111/j.0105-2896.2005.00301.x [DOI] [PubMed] [Google Scholar]
- 11.Shortman K, Heath WR. 2010. The CD8+ dendritic cell subset. Immunol. Rev. 234:18–31. 10.1111/j.0105-2896.2009.00870.x [DOI] [PubMed] [Google Scholar]
- 12.Kroczek RA, Henn V. 2012. The role of XCR1 and its ligand XCL1 in antigen cross-presentation by murine and human dendritic cells. Front. Immunol. 3:14. 10.3389/fimmu.2012.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2703–2772 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 14.Beisser PS, Lavreysen H, Bruggeman CA, Vink C. 2008. Chemokines and chemokine receptors encoded by cytomegaloviruses. Curr. Top. Microbiol. Immunol. 325:221–242. 10.1007/978-3-540-77349-8_13 [DOI] [PubMed] [Google Scholar]
- 15.Akter P, Cunningham C, McSharry BP, Dolan A, Addison C, Dargan DJ, Hassan-Walker AF, Emery VC, Griffiths PD, Wilkinson GW, Davison AJ. 2003. Two novel spliced genes in human cytomegalovirus. J. Gen. Virol. 84:1117–1122. 10.1099/vir.0.18952-0 [DOI] [PubMed] [Google Scholar]
- 16.Penfold ME, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. U. S. A. 96:9839–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lüttichau HR. 2010. The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J. Biol. Chem. 285:9137–9146. 10.1074/jbc.M109.002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straschewski S, Patrone M, Walther P, Gallina A, Mertens T, Frascaroli G. 2011. Protein pUL128 of human cytomegalovirus is necessary for monocyte infection and blocking of migration. J. Virol. 85:5150–5158. 10.1128/JVI.02100-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao H, Tao R, Zheng Q, Xu J, Shang S. 2013. Recombinant HCMV UL128 expression and functional identification of PBMC-attracting activity in vitro. Arch. Virol. 158:173–177. 10.1007/s00705-012-1378-8 [DOI] [PubMed] [Google Scholar]
- 20.Vomaske J, Denton M, Kreklywich C, Andoh T, Osborn JM, Chen D, Messaoudi I, Orloff SL, Streblow DN. 2012. Cytomegalovirus CC chemokine promotes immune cell migration. J. Virol. 86:11833–11844. 10.1128/JVI.00452-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Cleef KW, Smit MJ, Bruggeman CA, Vink C. 2006. Cytomegalovirus-encoded homologs of G protein-coupled receptors and chemokines. J. Clin. Virol. 35:343–348. 10.1016/j.jcv.2005.10.013 [DOI] [PubMed] [Google Scholar]
- 22.Vink C, Beuken E, Bruggeman CA. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saederup N, Aguirre SA, Sparer TE, Bouley DM, Mocarski ES. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J. Virol. 75:9966–9976. 10.1128/JVI.75.20.9966-9976.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voigt S, Sandford GR, Hayward GS, Burns WH. 2005. The English strain of rat cytomegalovirus (CMV) contains a novel captured CD200 (vOX2) gene and a spliced CC chemokine upstream from the major immediate-early region: further evidence for a separate evolutionary lineage from that of rat CMV Maastricht. J. Gen. Virol. 86:263–274. 10.1099/vir.0.80539-0 [DOI] [PubMed] [Google Scholar]
- 25.Noda S, Aguirre SA, Bitmansour A, Brown JM, Sparer TE, Huang J, Mocarski ES. 2006. Cytomegalovirus MCK-2 controls mobilization and recruitment of myeloid progenitor cells to facilitate dissemination. Blood 107:30–38. 10.1182/blood-2005-05-1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daley-Bauer LP, Wynn GM, Mocarski ES. 2012. Cytomegalovirus impairs antiviral CD8+ T cell immunity by recruiting inflammatory monocytes. Immunity 37:122–133. 10.1016/j.immuni.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ettinger J, Geyer H, Nitsche A, Zimmermann A, Brune W, Sandford GR, Hayward GS, Voigt S. 2012. Complete genome sequence of the english isolate of rat cytomegalovirus (Murid herpesvirus 8). J. Virol. 86:13838. 10.1128/JVI.02614-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallny HJ. 1997. Production of soluble MHC class II molecules in Drosophila melanogaster Schneider cells. Immunol. Methods Manual 1:51–59 [Google Scholar]
- 29.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 [DOI] [PubMed] [Google Scholar]
- 30.Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. 2003. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 376:952–965. 10.1007/s00216-003-2057-0 [DOI] [PubMed] [Google Scholar]
- 31.Bachem A, Güttler S, Hartung E, Ebstein F, Schaefer M, Tannert A, Salama A, Movassaghi K, Opitz C, Mages HW, Henn V, Kloetzel PM, Gurka S, Kroczek RA. 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281. 10.1084/jem.20100348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hautamaa D, Merica R, Chen Z, Jenkins MK. 1997. Murine lymphotactin: gene structure, post-translational modification and inhibition of expression by CD28 costimulation. Cytokine 9:375–382 [DOI] [PubMed] [Google Scholar]
- 33.Mortier A, Van Damme J, Proost P. 2008. Regulation of chemokine activity by posttranslational modification. Pharmacol. Ther. 120:197–217. 10.1016/j.pharmthera.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 34.Voisine C, Hubert FX, Trinite B, Heslan M, Josien R. 2002. Two phenotypically distinct subsets of spleen dendritic cells in rats exhibit different cytokine production and T cell stimulatory activity. J. Immunol. 169:2284–2291 http://www.jimmunol.org/content/169/5/2284.full [DOI] [PubMed] [Google Scholar]
- 35.Thelen M. 2001. Dancing to the tune of chemokines. Nat. Immunol. 2:129–134. 10.1038/84224 [DOI] [PubMed] [Google Scholar]
- 36.Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Güttler S, Hutloff A, Mages HW, Ranke K, Schaefer M, Jack RS, Henn V, Kroczek RA. 2009. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31:823–833. 10.1016/j.immuni.2009.08.027 [DOI] [PubMed] [Google Scholar]
- 37.Wang D, Bresnahan W, Shenk T. 2004. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. U. S. A. 101:16642–16647. 10.1073/pnas.0407233101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcami A, Saraiva M. 2009. Chemokine binding proteins encoded by pathogens. Adv. Exp. Med. Biol. 666:167–179. 10.1007/978-1-4419-1601-3_13 [DOI] [PubMed] [Google Scholar]
- 39.Murphy PM. 2001. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol. 2:116–122. 10.1038/84214 [DOI] [PubMed] [Google Scholar]
- 40.Kelner GS, Kennedy J, Bacon KB, Kleyensteuber S, Largaespada DA, Jenkins NA, Copeland NG, Bazan JF, Moore KW, Schall TJ, Zlotnik A. 1994. Lymphotactin: a cytokine that represents a new class of chemokine. Science 266:1395–1399 [DOI] [PubMed] [Google Scholar]
- 41.Loewendorf A, Benedict CA. 2010. Modulation of host innate and adaptive immune defenses by cytomegalovirus: timing is everything. J. Intern. Med. 267:483–501. 10.1111/j.1365-2796.2010.02220.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen SJ, Crown SE, Handel TM. 2007. Chemokine: receptor structure, interactions, and antagonism. Annu. Rev. Immunol. 25:787–820. 10.1146/annurev.immunol.24.021605.090529 [DOI] [PubMed] [Google Scholar]
- 43.Dorner BG, Steinbach S, Hüser MB, Kroczek RA, Scheffold A. 2003. Single-cell analysis of the murine chemokines MIP-1alpha, MIP-1beta, RANTES and ATAC/lymphotactin by flow cytometry. J. Immunol. Methods 274:83–91. 10.1016/S0022-1759(02)00498-2 [DOI] [PubMed] [Google Scholar]
- 44.Hubert FX, Voisine C, Louvet C, Heslan JM, Ouabed A, Heslan M, Josien R. 2006. Differential pattern recognition receptor expression but stereotyped responsiveness in rat spleen dendritic cell subsets. J. Immunol. 177:1007–1016 http://www.jimmunol.org/content/177/2/1007 [DOI] [PubMed] [Google Scholar]
- 45.Yrlid U, Macpherson G. 2003. Phenotype and function of rat dendritic cell subsets. APMIS 111:756–765. 10.1034/j.1600-0463.2003.11107807.x [DOI] [PubMed] [Google Scholar]
- 46.Trinite B, Chauvin C, Peche H, Voisine C, Heslan M, Josien R. 2005. Immature CD4− CD103+ rat dendritic cells induce rapid caspase-independent apoptosis-like cell death in various tumor and nontumor cells and phagocytose their victims. J. Immunol. 175:2408–2417 http://www.jimmunol.org/content/175/4/2408.full [DOI] [PubMed] [Google Scholar]
- 47.Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E, Vu Manh TP, Baranek T, Storset AK, Marvel J, Boudinot P, Hosmalin A, Schwartz-Cornil I, Dalod M. 2010. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 207:1283–1292. 10.1084/jem.20100223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrews DM, Scalzo AA, Yokoyama WM, Smyth MJ, Degli-Esposti MA. 2003. Functional interactions between dendritic cells and NK cells during viral infection. Nat. Immunol. 4:175–181. 10.1038/ni880 [DOI] [PubMed] [Google Scholar]
- 49.Dalod M, Hamilton T, Salomon R, Salazar-Mather TP, Henry SC, Hamilton JD, Biron CA. 2003. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon alpha/beta. J. Exp. Med. 197:885–898. 10.1084/jem.20021522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grigoleit U, Riegler S, Einsele H, Laib Sampaio K, Jahn G, Hebart H, Brossart P, Frank F, Sinzger C. 2002. Human cytomegalovirus induces a direct inhibitory effect on antigen presentation by monocyte-derived immature dendritic cells. Br. J. Haematol. 119:189–198. 10.1046/j.1365-2141.2002.03798.x [DOI] [PubMed] [Google Scholar]
- 51.Kessler T, Reich M, Jahn G, Tolosa E, Beck A, Kalbacher H, Overkleeft H, Schempp S, Driessen C. 2008. Human cytomegalovirus infection interferes with major histocompatibility complex type II maturation and endocytic proteases in dendritic cells at multiple levels. J. Gen. Virol. 89:2427–2436. 10.1099/vir.0.2008/001610-0 [DOI] [PubMed] [Google Scholar]
- 52.Andrews DM, Andoniou CE, Granucci F, Ricciardi-Castagnoli P, Degli-Esposti MA. 2001. Infection of dendritic cells by murine cytomegalovirus induces functional paralysis. Nat. Immunol. 2:1077–1084. 10.1038/ni724 [DOI] [PubMed] [Google Scholar]
- 53.Varani S, Frascaroli G, Homman-Loudiyi M, Feld S, Landini MP, Soderberg-Naucler C. 2005. Human cytomegalovirus inhibits the migration of immature dendritic cells by down-regulating cell-surface CCR1 and CCR5. J. Leukoc. Biol. 77:219–228. 10.1189/jlb.0504301 [DOI] [PubMed] [Google Scholar]
- 54.Steinman RM, Hawiger D, Liu K, Bonifaz L, Bonnyay D, Mahnke K, Iyoda T, Ravetch J, Dhodapkar M, Inaba K, Nussenzweig M. 2003. Dendritic cell function in vivo during the steady state: a role in peripheral tolerance. Ann. N. Y. Acad. Sci. 987:15–25. 10.1111/j.1749-6632.2003.tb06029.x [DOI] [PubMed] [Google Scholar]
- 55.Wehner R, Dietze K, Bachmann M, Schmitz M. 2011. The bidirectional crosstalk between human dendritic cells and natural killer cells. J. Innate Immun. 3:258–263. 10.1159/000323923 [DOI] [PubMed] [Google Scholar]
- 56.Scalzo AA, Corbett AJ, Rawlinson WD, Scott GM, Degli-Esposti MA. 2007. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol. Cell Biol. 85:46–54. 10.1038/sj.icb.7100013 [DOI] [PubMed] [Google Scholar]
- 57.Biron CA, Byron KS, Sullivan JL. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320:1731–1735 [DOI] [PubMed] [Google Scholar]
- 58.Reddehase MJ, Weiland F, Münch K, Jonjic S, Lüske A, Koszinowski UH. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. 1992. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science 257:238–241 [DOI] [PubMed] [Google Scholar]
- 60.Kapp M, Tan SM, Einsele H, Grigoleit G. 2007. Adoptive immunotherapy of HCMV infection. Cytotherapy 9:699–711. 10.1080/14653240701656046 [DOI] [PubMed] [Google Scholar]
- 61.Sinclair J. 2008. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J. Clin. Virol. 41:180–185. 10.1016/j.jcv.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 62.Sinclair J, Sissons P. 2006. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 87:1763–1779. 10.1099/vir.0.81891-0 [DOI] [PubMed] [Google Scholar]
- 63.Kaptein SJ, van Cleef KW, Gruijthuijsen YK, Beuken EV, van Buggenhout L, Beisser PS, Stassen FR, Bruggeman CA, Vink C. 2004. The r131 gene of rat cytomegalovirus encodes a proinflammatory CC chemokine homolog which is essential for the production of infectious virus in the salivary glands. Virus Genes 29:43–61. 10.1023/B:VIRU.0000032788.53592.7c [DOI] [PubMed] [Google Scholar]
- 64.Lüttichau HR, Lewis IC, Gerstoft J, Schwartz TW. 2001. The herpesvirus 8-encoded chemokine vMIP-II, but not the poxvirus-encoded chemokine MC148, inhibits the CCR10 receptor. Eur. J. Immunol. 31:1217–1220. [DOI] [PubMed] [Google Scholar]
- 65.Lüttichau HR, Johnsen AH, Jurlander J, Rosenkilde MM, Schwartz TW. 2007. Kaposi sarcoma-associated herpes virus targets the lymphotactin receptor with both a broad spectrum antagonist vCCL2 and a highly selective and potent agonist vCCL3. J. Biol. Chem. 282:17794–17805. 10.1074/jbc.M702001200 [DOI] [PubMed] [Google Scholar]