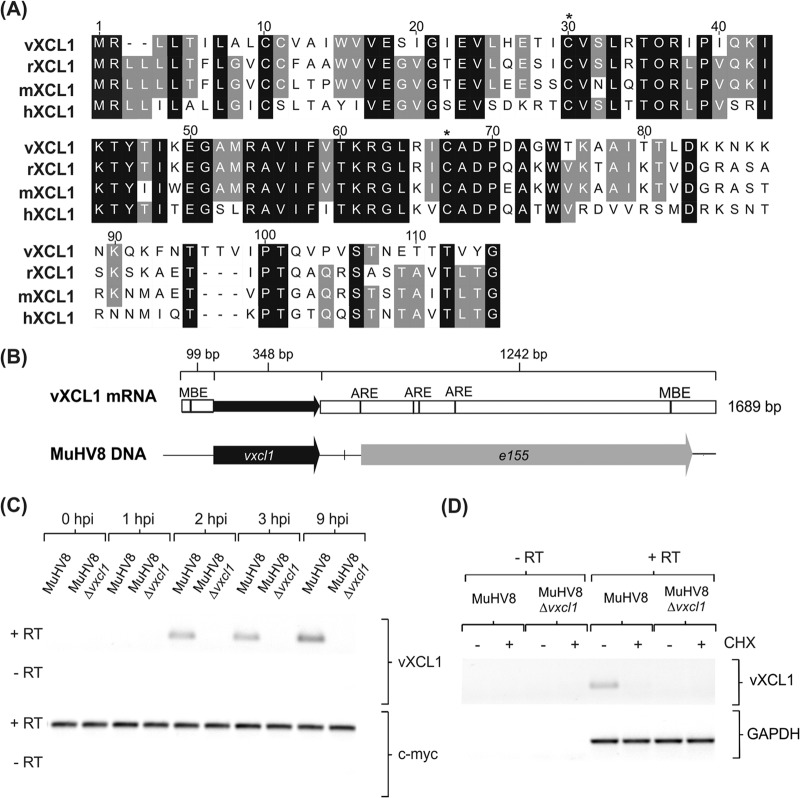
FIG 1.
Characterization of MuHV8-encoded C chemokine vXCL1. (A) Muscle alignment (Geneious version 5.5.7) of amino acid sequences of vXCL1 and rat, mouse, and human XCL1. Identical amino acids are indicated by a black background and similar amino acids by a gray background, and lack of similarity is indicated by a white background. Cysteine positions characteristic of C chemokines are indicated by asterisks. Numbers refer to the amino acid positions of the viral chemokine. Accession numbers for the sequences depicted are JX867617.1 (vXCL1, MuHV8), NP_599188 (rXCL1, Rattus norvegicus XCL1), NP_032536 (mXCL1, Mus musculus XCL1), and NP_002986 (hXCL1, Homo sapiens XCL1). (B) Analysis of vXCL1 mRNA. Total RNA from MuHV8-infected REF was isolated 24 h postinfection (hpi) and 3′- and 5′-UTRs were determined. Musashi binding elements (MBE) were detected using UTRScan, and AU-rich elemenst (ARE) were identified by scanning for an AUUUA element with CloneManager (Sci-Ed software, Cary, NC, USA). (C) Time course of MuHV8 vXCL1 expression. Total RNA from wild-type MuHV8 and MuHV8 Δvxcl1-infected cells was harvested at indicated time points, reverse transcribed with oligo(dT) primers, and amplified with vxcl1-specific primers. (D) Cycloheximide (CHX) was used to discriminate between immediate early and early gene expression. MuHV8 Δvxcl1 was included as a negative control. The experiment was carried out at least twice with similar results. c-myc and GAPDH served as controls. RT, reverse transcriptase.