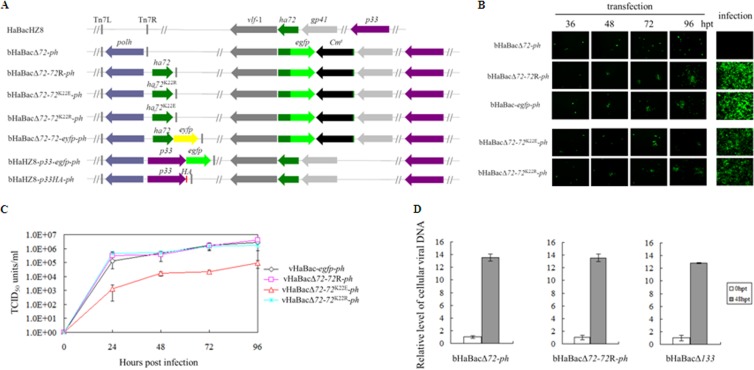
FIG 2.
HA72 is essential for BV production but not DNA replication, and the K22E mutation curtails BV production. (A) Schematic diagrams of recombinant bacmids. The ha72 ORF was partially replaced with an egfp and Cm resistance gene cassette from pKS-egfp-Cmr, which contains an egfp gene driven by an hsp70 promoter and a Cmr gene under the control of the T7 promoter, to generate ha72-inactivated bacmid bHaBacΔ72. The recombinant bacmids bHaBacΔ72-ph, bHaBacΔ72-72R-ph, bHaBacΔ72-72K22E-ph, bHaBacΔ72-72K22R-ph, and bHaBacΔ72-72-eyfp-ph were generated by Tn7-based transposition of the indicated genes into the ph locus of bHaBacΔ72. bHaBacΔ72-p33-egfp-ph and bHaBacHZ8-p33HA-ph were generated by Tn7-based transposition of the indicated genes into bHaBacHZ8. (B) Analysis of virus replication and BV production in HzAM1 cells. Fluorescence microscopy images were taken at various time posttransfection or at 120 hpi. (C) One-step growth curve analysis of recombinant viruses. HzAM1 cells were infected with vHaBac-egfp-ph, vHaBacΔ72-72R-ph, vHaBacΔ72-72K22E-ph, or vHaBacΔ72-72K22R-ph at a multiplicity of infection of 5. Supernatants were harvested at the designated time points postinfection and quantified for production of infectious BV by the endpoint dilution method. Data points represent average titers from triplicate infections. Error bars represent standard errors. (D) Real-time PCR analysis of viral DNA replication. HzAM1 cells were transfected with 3 μg bHaBacΔ72-ph, bHaBacΔ72-72R-ph, or control bacmid, bHaBacΔ133 (null for the f protein gene) through three experiments with three replicates per experiment. At the indicated time points postinfection, cells were collected to extract total DNA. Prior to quantitative PCR, DNA was digested with DpnI to avoid interference from E. coli-derived viral DNA. Quantitative PCRs were carried out in triplicates, utilizing 1 μl of DpnI-treated DNA. Error bars represent the standard errors.