Abstract
Introduction
Previous research suggests that aspects of facial surface morphology are heritable. Traditionally, heritability studies have used a limited set of linear distances to quantify facial morphology and often employ statistical methods poorly designed to deal with biological shape. In this preliminary report, we use a combination of 3D photogrammetry and landmark-based morphometrics to explore which aspects of face shape show the strongest evidence of heritability in a sample of twins.
Methods
3D surface images were obtained from 21 twin pairs (10 monozygotic, 11 same-sex dizygotic). Thirteen 3D landmarks were collected from each facial surface and their coordinates subjected to geometric morphometric analysis. This involved superimposing the individual landmark configurations and then subjecting the resulting shape coordinates to a principal components analysis. The resulting PC scores were then used to calculate rough narrow-sense heritability estimates.
Results
Three principal components displayed evidence of moderate to high heritability and were associated with variation in the breadth of orbital and nasal structures, upper lip height and projection, and the vertical and forward projection of the root of the nose due to variation in the position of nasion.
Conclusions
Aspects of facial shape, primarily related to variation in length and breadth of central midfacial structures, were shown to demonstrate evidence of strong heritability. An improved understanding of which facial features are under strong genetic control is an important step in the identification of specific genes that underlie normal facial variation.
Keywords: 3D stereophotogrammetry, face shape, geometric morphometrics, heritability, twins
Introduction
Variation in human facial morphology is strongly influenced by genetics. This contention is supported by decades of accumulated anthropometric research on twin concordance and parent-offspring resemblance [1-16]. To summarize the major findings of the previous research: 1) estimates of heritability for individual or composite quantitative facial features generally range from moderate (0.40) to high (0.80); 2) additive genetic effects account for much of the observed phenotypic variance in facial traits; 3) measured variables in each of the principal anatomical directions (horizontal, vertical and projective) display strong evidence of heritability; 4) vertically oriented measures (e.g., facial height measures) tend to be among the most heritable facial traits. Determining the genetic contribution for specific facial traits remains a relevant area of concern in a number of fields that deal with human morphology, including paleoanthropology and orthodontics.
While the majority of craniofacial heritability studies are limited to measures of skeletal structure, most often derived from radiographs, a smaller number of studies have focused solely on external soft-tissue features [17-35]. These studies typically use traditional anthropometric methods (i.e., direct measurement with calipers) to quantify the head and face, reporting similar heritability estimates for these surface traits as for the underlying skull. 3D surface imaging represents an alternative, and far less invasive, method for capturing quantitative data on the facial soft tissues [36,37]. Recent advances in capture speed, accuracy, and affordability have rendered traditional methods of direct facial measurement largely obsolete. Moreover, by combining 3D surface imaging technology with powerful methods of landmark-based shape analysis, it is possible to quantify subtle aspects of facial morphology and variation often missed with conventional anthropometrics. Nevertheless, only two published studies to date have utilized 3D surface imaging to evaluate the genetic basis of facial features [38,39]. Although both studies found a very high degree of similarity among monozygotic twins compared with same-sex dizygotic pairs, with the central midfacial region exhibiting the greatest level of concordance, the quantitative analysis was limited to a small set of simple linear distances, which are often inadequate for describing complex 3D features.
In this preliminary investigation, we utilize 3D digital stereophotogrammetry in concert with geometric morphometrics to quantify midfacial structures in a sample of monozygotic (MZ) and same-sex dizygotic (DZ) twins. The main objective is to identify aspects of midfacial shape that show the strongest evidence of heritability.
Materials and Methods
A set of 10 monozygotic twin pairs (20 individuals) and 11 same-sex dizygotic twin pairs (22 individuals) were ascertained at the 2005 annual Twins Days Festival in Twinsburg, Ohio. The twin sample was comprised of Caucasian boys and girls between the ages of 5 and 12 years (mean age = 9.3 years ±2.2). Following informed consent, a 3D facial photo was obtained from each child as part of a broader assessment of phenotypic traits. Medical history was also collected from all participants to rule out any congenital conditions or trauma that might affect facial morphology. Routine orthodontic interventions were not considered as a basis for exclusion. Parents of same-sex twins completed a standard 15-item questionnaire regarding the twins similarities and differences to determine zygosity [40]. The algorithm for determining zygosity has been described elsewhere.41 It has been previously demonstrated that this short questionnaire has an accuracy of 94% for zygosity determination when compared with molecular genetic results [41,42]. All aspects of this study were approved by an Institutional Review Board (IRB).
Facial surface images were obtained via 3D digital stereophotogrammetry (FaceCam 250, Genex Technologies, Kensington, MD). Briefly, the imaging process involves projecting a structured light pattern onto the facial surface, which is then recorded by multiple CCD cameras with overlapping fields of view. By extracting positional information from the deformation of the projected light pattern by the natural contours of the target surface, the location of points (> 300,000) distributed over the entire surface is determined. From this collection of points, an accurate 3D model of the facial surface is then generated (Fig. 1). Color and texture information is captured simultaneously and is mapped onto the underlying surface geometry, providing lifelike rendering. The entire image acquisition process takes place in less than 0.5 seconds. The accuracy and precision of measures derived these 3D surface models has been explored previously by the authors and found to very high [43,44].
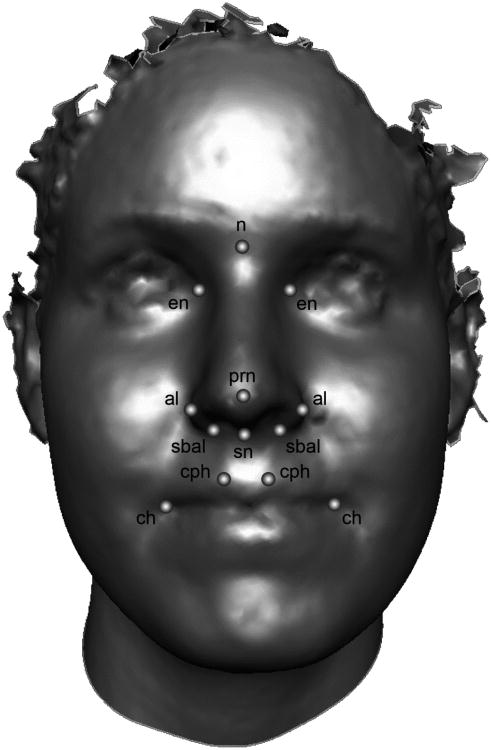
Fig 1.
3D facial surface showing the midfacial landmarks used in the present study. Midline points: n = nasion, prn = pronasale, sn = subnasale. Bilateral points: en = endocanthion, al = alare, sbal = subalare, cph = crista philtri, ch = chelion.
Thirteen surface landmarks were collected from each 3D facial model (Fig. 1) and their XYZ coordinate locations saved. These landmarks correspond primarily to centrally located midfacial structures and are defined as in traditional surface anthropometry [45]. There were several factors that limited the number and kinds of landmarks we were able to collect. For many subjects the quality of the scans was compromised below the labial fissure, eliminating the opportunity to capture landmarks on the lower lips or chin. Furthermore, a number of subjects failed to maintain open eyes during the capture, making the identification of exocanthion impossible. A geometric morphometric approach was utilized to perform shape analysis of the landmark coordinate data [46,47]. First, the individual landmark configurations for all 42 participants were scaled and aligned though a least-squares Procrustes superimposition [48]. This procedure results in a new set of 3D coordinates (Procrustes coordinates), preserving only shape information. To control for any remaining allometry effects in the data, centroid size was regressed against the Procrustes shape coordinates, and the resulting residuals were saved for further analysis. Principal components analysis (PCA) was then applied to the allometry-adjusted Procrustes coordinates in order to describe the shape variation present in the dataset. In the context of shape analysis, PCA reduces the dimensionality of the Procrustes coordinate data into a more manageable number of uncorrelated summary variables (components), each capturing a distinct mode of shape variation [49]. Because the geometric morphometric approach preserves the intrinsic geometry present in landmark coordinate data, shape variation associated with a given principal component (PC) can be visualized intuitively as a displacement of points in 3D space [50,51]. To further enhance visualization, facial surface warps can be generated based on the PCA results.
Similar to traditional PCA or factor analysis, every subject in the dataset receives a score on each extracted PC. These PC scores can then be used to compute narrow-sense heritability (G) estimates with the following formula:
where ICC is the intraclass correlation coefficient. This is a very crude method for estimating heritability, often used when sample sizes preclude the use of more refined methods, such as structural equation modeling. All geometric morphometric analyses (Procrustes analysis, allometry regression, and PCA) were carried out using the program MorphoJ [52]. Facial surface warps based on the PCA results were generated using the program Landmark v3.6 (http://www.idav.ucdavis.edu/research/EvoMorph).
Results
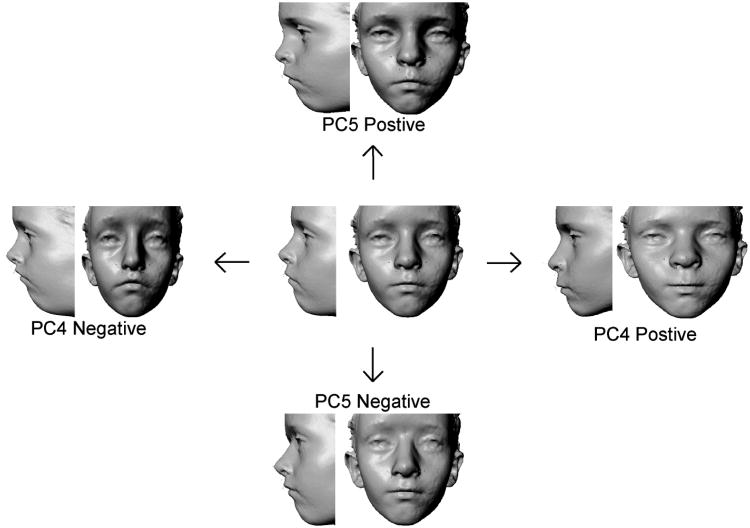
A total of 17 PCs were extracted; ICCs were only calculated from scores on the first 9 PCs, which accounted for approximately 90% of the total shape variance. Three of the derived shape PCs displayed evidence of moderate to high heritability (PC4, PC5 and PC7); the remaining six PCs were unrevealing. The heritability estimates for PC4, PC5 and PC7 are provided in Table 1; although the values reported here approach 1.0, it should be noted these are still very crude approximations. We attempted to use bootstrap resampling techniques to obtain empirical confidence intervals for these estimates. These resulting confidence intervals approached or included zero and were likely affected by the small sample size from which the permutations were based. Individually these PCs accounted for a relatively small amount of the total shape variance (PC4 = 6.8%, PC5 = 5.7%, PC7 = 4.0%). PC4 was associated with a complex suite of shape variations. In the upper face, variation in the lateral position of the left/right endocanthion points (relating to the degree of hyper/hypotelorism) was observed. Variation in nasal breadth, height and projection was due primarily to shifts in the vertical and anterior-posterior position of nasion, the lateral position of the alar cartilages (left/right alare), and the anterior-posterior position of the nasal tip (pronasale). In the orolabial region, PC4 was associated with variation in the length of the philtrum and vertical height of the upper lip; this was coupled with relative changes in the vertical position of the left and right chelion. PC5 was associated mainly the vertical and anterior-posterior position of nasion, the anterior-posterior position of the orbits (left/right endocanthion), and the breadth of the nasal floor and philtrum. Variation in shape associated with PC4 and PC5 are depicted as surface warps in Figure 2. PC7 (not shown in Figure 2) was related primarily to the vertical position of landmarks defining the interorbital septum (nasion and the left/right endocanthion) and variation in the nasolabial angle due to the anterior position of the upper lip relative to the nose. The first few PCs, accounting for the majority of shape variation in the sample did not demonstrate strong evidence of heritability. Instead, shape variation along these PCs was related more to sex and age/maturity related factors. The source of the variation for the remaining PCs was not immediately apparent.
Table 1.
Intraclass correlation coefficients (ICC) and G estimates for principal components showing evidence of high heritability.
| Shape Component | Variation Explained | ICCMZ | ICCDZ | Ga |
|---|---|---|---|---|
| PC4 | 6.8% | 0.783* | 0.013 | 1.0b |
| PC5 | 5.7% | 0.771* | 0.314 | 0.9 |
| PC7 | 4.0% | 0.675* | 0.284 | 1.0b |
p < 0.05 for test of within group non-zero ICC
G = 2(ICCMZ − ICCDZ)
Heritability estimate capped at 1.0
Fig 2.
Facial surface warps modeling the shape variation associated with PC4 (along the horizontal axis) and PC5 (along the vertical axis). The centrally located face represents the average or consensus facial surface for the sample. Movement along the principal axes of shape variation in either direction away from the consensus face is accompanied by the 3D displacement of landmarks and modeled as a deformation of the consensus 3D surface. This deformation reflects the major modes of shape variation associated with PC4 and PC5, respectively.
Discussion
The results of the present study revealed evidence of heritable shape variation in central midfacial structures, including the interorbital region, nose and upper lip. Principal shape components associated with the degree of horizontal separation between the eyes, the length, breadth and projection of the nose, and the height and projection of the upper lip all demonstrated high heritability. These findings are largely supported by more traditional anthropometric studies that utilize standard linear distances to quantify facial surface structures [18-20,22-35]. There is also substantial overlap between the results of the present study and those of prior 3D facial surface studies on twin pairs. Burke [38] employed an early form of stereophotogrammetry to assess facial similarity in a sample of 18 like-sexed twin pairs. Of the 13 facial measures examined, the degree of intrapair difference was significantly greater in DZ compared to MZ twins for eight variables, most of which involved central midfacial structures including the nose, interorbital region and upper lip. Using a similar study design, Naini and Moss [39] reported comparable results in a sample of 20 like-sexed twin pairs using 3D laser scanning technology. Kau et al. [53] utilized 3D laser surface scanning to evaluate facial resemblance in a single pair of monozygotic twins; within-pair differences at points spread over the facial surface were generally at the sub-millimeter level (the chin region showed the greatest disparity at 1.5-1.9mm).
Several prior studies report the highest heritability estimates for vertically oriented facial measures [6,9-11,24,54], although there is considerable disagreement on this point [7,14,33-35]. The current findings present a slightly more complex picture. PC4 was associated with simultaneous changes in the relative length and breadth of the midface. This pattern of correlated shifts in length and breadth contributed to an overall shift in midface shape, from narrow and long to short and broad (see Figure 2). The remaining two PCs showing evidence of heritability, however, were related primarily to the vertical and anterior position of the midline landmark nasion. Thus, there is only partial support for the contention that vertical aspects of the face show higher heritability relative to variation along other principal anatomical directions.
Determining the heritability of quantitative facial traits represents an important step in the effort to identify specific genes that underlie typical variation in face shape. To date, limited work in this area has been carried out on animal models. Quantitative trait loci (QTL) have been identified in mice that relate to variation in mandibular [55-57] and maxillary shape [58,59]. In particular, QTLs on several chromosomes have been associated with variation in midfacial breadth and length (projection) in the mouse skull [58,59]. In dogs, polymorphisms in specific candidate genes (e.g., Tcof1, Fgf8, Runx2) have been shown to influence the extreme variation in facial form that characterizes certain breeds [60-63]. In humans, a handful of recent studies have also begun to uncover the genetic basis of facial shape [64,65]. Interestingly, both of the aforementioned human studies have independently identified associations between SNP variants in the PAX3 gene and variation in nasal root morphology, specifically relating to the relative position of nasion and left/right endocanthion. Variation in the spatial position of these same points was associated with all three relevant PCs in the present study, suggesting that our preliminary results, although quite limited, may have some external validity.
It is important to keep in mind that although several derived shape PCs showed seemingly very high levels of heritability, these estimates are based on a simplistic approach to modeling heritability in a small sample and ultimately could not be substantiated using resampling methods. Consequently, the heritability values for PC4, PC5 and PC7 are likely to be overestimates of the true additive genetic effects on phenotypic similarity. Rather than focusing on the values in an absolute sense, a more prudent interpretation might be that PC4, PC5 and PC7 demonstrated the most compelling evidence of heritability, relative to the other components of shape variation derived here. In future studies, larger samples will hopefully permit the use of more sophisticated methods for estimating heritability, including those based on structural equation modeling.
Conclusion
This study represents a preliminary attempt to estimate the heritability of facial shape using a combination of 3D stereophotogrammetry and landmark-based geometric morphometrics in a sample of twin pairs. Our results indicate that variation in both the length and breadth of central midfacial structures showed statistical evidence of moderate to high heritability. These results are in line with previous reports, including recent studies that have identified specific genetic variants underlying facial shape in humans. Nevertheless, due to the small size of the sample of twin pairs available for analysis, the results here should be viewed as preliminary and, as such, interpreted with caution.
Acknowledgments
The authors would like to thank Carla Brandon and Kathy Bardi for their help recruiting subjects and Nicole Scott for helping to collect 3D images. We also thank the directors of the Twins Days Festival for granting permission to conduct this research. This work was supported by the following grant from the National Institute of Dental and Craniofacial Research: R01-DE016148. The authors have no conflict of interest to declare.
References
- 1.Nakata M, Yu PL, Davis B, Nance WE. Genetic determinants of cranio-facial morphology: a twin study. Ann Hum Genet. 1974;37:431–43. doi: 10.1111/j.1469-1809.1974.tb01848.x. [DOI] [PubMed] [Google Scholar]
- 2.Nakata M, Yu PL, Nance WE. Multivariate analysis of craniofacial measurements in twin and family data. Am J Phys Anthropol. 1974;41:423–9. doi: 10.1002/ajpa.1330410309. [DOI] [PubMed] [Google Scholar]
- 3.Saunders SR, Popovich F, Thompson GW. A family study of craniofacial dimensions in the Burlington Growth Center sample. Am J Orthod. 1980;78:394–403. doi: 10.1016/0002-9416(80)90020-2. [DOI] [PubMed] [Google Scholar]
- 4.Byard PJ, Lewis AB, Ohtsuki F, Siervogel RM, Roche AF. Sibling correlations for cranial measurements from serial radiographs. J Craniofac Genet Dev Biol. 1984;4:265–9. [PubMed] [Google Scholar]
- 5.Lobb WK. Craniofacial morphology and occlusal variation in monozygotic and dizygotic twins. Angle Orthodont. 1987;57:219–33. doi: 10.1043/0003-3219(1987)057<0219:CMAOVI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 6.Lundstrom A, McWilliam J. Comparison of some cephalometric distances and corresponding facial proportions with regard to heritability. Eur J Orthod. 1988;10:27–9. doi: 10.1093/ejo/10.1.27. [DOI] [PubMed] [Google Scholar]
- 7.Harris EF, Johnson MG. Heritability of craniometric and occlusal variables: a longitudinal sib analysis. Am J Orthod Dentofacial Orthop. 1991;99:258–68. doi: 10.1016/0889-5406(91)70007-J. [DOI] [PubMed] [Google Scholar]
- 8.King L, Harris EF, Tolley EA. Heritability of cephalometric and occlusal variables as assessed from siblings with overt malocclusions. Am J Orthod Dentofacial Orthop. 1993;104:121–31. doi: 10.1016/S0889-5406(05)81001-7. [DOI] [PubMed] [Google Scholar]
- 9.Manfredi C, Martina R, Grossi GB, Giuliani M. Heritability of 39 orthodontic cephalometric parameters on MZ, DZ twins and MN-paired singletons. Am J Orthod Dentofacial Orthop. 1997;111:44–51. doi: 10.1016/s0889-5406(97)70301-9. [DOI] [PubMed] [Google Scholar]
- 10.Savoye I, Loos R, Carels C, Derom C, Vlietinck R. A genetic study of anteroposterior and vertical facial proportions using model-fitting. Angle Orthodont. 1998;68:467–70. doi: 10.1043/0003-3219(1998)068<0467:AGSOAA>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Carels C, Van Cauwenberghe N, Savoye I, et al. A quantitative genetic study of cephalometric variables in twins. Orthod Craniofacial Res. 2001;4:130–40. doi: 10.1034/j.1600-0544.2001.040303.x. [DOI] [PubMed] [Google Scholar]
- 12.Johannsdottir B, Thorarinsson F, Thordarson A, Magnusson TE. Heritability of craniofacial characteristics between parents and offspring from lateral cephalograms. Am J Orthod Dentofacial Orthop. 2005;127:200–7. doi: 10.1016/j.ajodo.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 13.Carson EA. Maximum likelihood estimation of human craniometric heritabilities. Am J Phys Anthropol. 2006;131:169–80. doi: 10.1002/ajpa.20424. [DOI] [PubMed] [Google Scholar]
- 14.Baydaş B, Erdem A, Yavuz I, Ceylan I. Heritability of facial proportions and soft-tissue profile characteristics in Turkish Anatolian siblings. Am J Orthod Dentofacial Orthop. 2007;131:504–9. doi: 10.1016/j.ajodo.2005.05.055. [DOI] [PubMed] [Google Scholar]
- 15.Sherwood RJ, Duren DL, Demerath EW, Czerwinski SA, Siervogel RM, Towne B. Quantitative genetics of modern human cranial variation. J Hum Evol. 2008;54:909–14. doi: 10.1016/j.jhevol.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Abadías N, Esparza M, Sjøvold T, González-José R, Santos M, Hernández M. Heritability of human cranial dimensions: comparing the evolvability of different cranial regions. J Anat. 2009;214:19–35. doi: 10.1111/j.1469-7580.2008.01015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howells WW. Correlation of brothers in factor scores. Am J Phys Anthropol. 1953;11:121–40. doi: 10.1002/ajpa.1330110119. [DOI] [PubMed] [Google Scholar]
- 18.Clark PJ. The heritability of certain anthropometric characters as ascertained from measurements of twins. Am J Hum Genet. 1956;8:49–54. [PMC free article] [PubMed] [Google Scholar]
- 19.Osborne RH, De George FV. Genetic Basis of Morphological Variation. Cambridge: Harvard University Press; 1959. [Google Scholar]
- 20.Vandenberg SG, Strandskov HH. A comparison of identical and fraternal twins on some anthropometric measures. Hum Biol. 1964;36:45–52. [PubMed] [Google Scholar]
- 21.Susanne C. Genetic and environmental influences on morphological characteristics. Ann Hum Biol. 1975;2:279–87. doi: 10.1080/03014467500000851. [DOI] [PubMed] [Google Scholar]
- 22.Susanne C. Heritability of anthropometric characters. Hum Biol. 1977;49:573–80. [PubMed] [Google Scholar]
- 23.Byard PJ, Sharma K, Russell JM, Rao DC. A family study of anthropometric traits in a Punjabi community: II. An investigation of familial transmission. Am J Phys Anthropol. 1984;64:97–104. doi: 10.1002/ajpa.1330640202. [DOI] [PubMed] [Google Scholar]
- 24.Byard PJ, Poosha DVR, Satyanarayana M, Rao DC. Family resemblance for components of craniofacial size and shape. J Craniofac Genet Dev Biol. 1985;5:229–38. [PubMed] [Google Scholar]
- 25.Poosha DV, Byard PJ, Satyanarayana M, Rice JP, Rao DC. Family resemblance for cranio- facial measurements in Velanti Brahmins from Andhra Pradesh, India. Am J Phys Anthropol. 1984;65:15–22. doi: 10.1002/ajpa.1330650104. [DOI] [PubMed] [Google Scholar]
- 26.Sharma K, Byard PJ, Russell JM, Rao DC. A family study of anthropometric traits in a Punjabi community: I. Introduction and familial correlations. Am J Phys Anthropol. 1984;63:389–95. doi: 10.1002/ajpa.1330630406. [DOI] [PubMed] [Google Scholar]
- 27.Hauspie RC, Susanne C, Defrise-Gussenhoven E. Testing for the presence of genetic variance in factors of face measurements of Belgian twins. Ann Hum Biol. 1985;12:429–40. doi: 10.1080/03014468500007991. [DOI] [PubMed] [Google Scholar]
- 28.Devor EJ, McGue M, Crawford MH, Lin PM. Transmissible and nontransmissible components of anthropometric variation in the Alexanderwohl Mennonites: I. Description and familial correlations. Am J Phys Anthropol. 1986;69:71–82. doi: 10.1002/ajpa.1330690109. [DOI] [PubMed] [Google Scholar]
- 29.Devor EJ, McGue M, Crawford MH, Lin PM. Transmissible and nontransmissible components of anthropometric variation in the Alexanderwohl Mennonites: II. Resolution by path analysis. Am J Phys Anthropol. 1986;69:83–92. doi: 10.1002/ajpa.1330690110. [DOI] [PubMed] [Google Scholar]
- 30.Devor EJ. Transmission of human craniofacial dimensions. J Craniofac Genet Dev Biol. 1987;7:95–106. [PubMed] [Google Scholar]
- 31.Livshits G, Otremski I, Kobyliansky E. Genetics of human body size and shape: complex segregation analysis. Ann Hum Biol. 1995;22:13–27. doi: 10.1080/03014469500003672. [DOI] [PubMed] [Google Scholar]
- 32.Nikolova M. Similarities in anthropometrical traits of children and their parents in a Bulgarian population. Ann Hum Genet. 1996;60:517–25. doi: 10.1111/j.1469-1809.1996.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 33.Arya R, Duggirala R, Comuzzie AG, Puppala S, Modem S, Busi BR, Crawford MH. Heritability of anthropometric phenotypes in caste populations of Visakhapatnam, India. Hum Biol. 2002;74:325–44. doi: 10.1353/hub.2002.0026. [DOI] [PubMed] [Google Scholar]
- 34.Ermakov S, Kobyliansky E, Livshits G. Quantitative genetic study of head size related phenotypes in ethnically homogeneous Chuvasha pedigrees. Ann Hum Biol. 2005;32:585–98. doi: 10.1080/03014460500247972. [DOI] [PubMed] [Google Scholar]
- 35.Karmakar B, Ermakov S, Yakovenko K, Kobyliansky E. Genetic determination of head-size- related anthropometric traits in an ethnically homogeneous sample of 373 Indian pedigrees of West Bengal. Hum Biol. 2007;79:501–14. doi: 10.1353/hub.2008.0007. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg SM, Kolar JC. Three-dimensional surface imaging: limitations and considerations from the anthropometric perspective. J Craniofac Surg. 2005;16:847–51. doi: 10.1097/01.scs.0000164330.96675.0c. [DOI] [PubMed] [Google Scholar]
- 37.Kau CH, Richmond S, Savio C, Mallorie C. Measuring adult facial morphology in three dimensions. Angle Orthodont. 2006;76:771–6. doi: 10.1043/0003-3219(2006)076[0773:MAFMIT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 38.Burke PH. Intrapair facial differences in twins. Acta Genet Med Gemellol (Roma) 1989;38:37–47. doi: 10.1017/s0001566000002816. [DOI] [PubMed] [Google Scholar]
- 39.Naini FB, Moss JP. Three-dimensional assessment of the relative contribution of genetics and environment to various facial parameters with the twin method. Am J Orthod Dentofacial Orthop. 2004;126:655–65. doi: 10.1016/j.ajodo.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 40.Nichols RC, Bilbro WC. Diagnosis of twin zygosity. Acta Genet Stat Med. 1966;16:265–75. doi: 10.1159/000151973. [DOI] [PubMed] [Google Scholar]
- 41.Strassberg M, Peters K, Marazita M, et al. Pittsburgh Registry of Infant Multiplets (PRIM) Twin Res. 2002;5:499–501. doi: 10.1375/136905202320906363. [DOI] [PubMed] [Google Scholar]
- 42.Rowe DC. Environmental and genetics influence on dimensions of perceived parenting: a twin study. Dev Psychol. 1981;17:203–8. [Google Scholar]
- 43.Weinberg SM, Scott NM, Neiswanger K, Brandon CA, Marazita ML. Digital three- dimensional photogrammetry: evaluation of anthropometric precision and accuracy using a Genex 3D camera system. Cleft Palate Craniofac J. 2004;41:507–18. doi: 10.1597/03-066.1. [DOI] [PubMed] [Google Scholar]
- 44.Weinberg SM, Naidoo S, Govier DP, Martin RA, Kane AA, Marazita ML. Anthropometric precision and accuracy of digital three-dimensional photogrammetry: comparing the Genex and 3dMD imaging systems to one another and to direct anthropometry. J Craniofac Surg. 2006;17:477–83. doi: 10.1097/00001665-200605000-00015. [DOI] [PubMed] [Google Scholar]
- 45.Farkas LG. Anthropometry of the Head and Face. New York: Raven Press; 1994. [Google Scholar]
- 46.Dryden IL, Mardia KV. Statistical Shape Analysis. Chichester: Wiley; 1998. [Google Scholar]
- 47.Slice DE. Modern morphometrics. In: Slice DE, editor. Modern Morphometrics in Physical Anthropology. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 1–45. [Google Scholar]
- 48.Rohlf F, Slice D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Zool. 1990;39:40–59. [Google Scholar]
- 49.Zelditch ML, Swiderski DL, Sheets HD, Fink WL. Geometric Morphometrics for Biologists: A Primer. Amsterdam: Elsevier Academic Press; 2004. [Google Scholar]
- 50.O'Higgins P, Jones N. Facial growth in Cercocebus torquantus: an application of three-dimensional geometric morphometric techniques to the study of morphological variation. J Anat. 1998;193:251–72. doi: 10.1046/j.1469-7580.1998.19320251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hennessy RJ, Stringer CB. Geometric morphometric study of the regional variation of modern human craniofacial form. Am J Phys Anthropol. 2002;117:37–48. doi: 10.1002/ajpa.10005. [DOI] [PubMed] [Google Scholar]
- 52.Klingenberg CP. MorphoJ: an integrated software package for geometric morphometrics. Mol Ecol Resour. 2011;11:353–7. doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- 53.Kau CH, Zhurov A, Bibb R, Hunter L, Richmond S. The investigation of the changing facial appearance of identical twins employing a three-dimensional laser imaging system. Orthod Craniofacial Res. 2005;8:85–90. doi: 10.1111/j.1601-6343.2005.00320.x. [DOI] [PubMed] [Google Scholar]
- 54.Hunter WS. A study of the inheritance of craniofacial characteristics as seen in lateral cephalograms of 72 like-sexed twins. Rep Congr Eur Orthod Soc. 1965;41:59–70. [PubMed] [Google Scholar]
- 55.Cheverud JM, Routman EJ, Irschick DJ. Pleiotropic effects of individual gene loci on mandibular morphology. Evolution. 1997;51:2006–16. doi: 10.1111/j.1558-5646.1997.tb05122.x. [DOI] [PubMed] [Google Scholar]
- 56.Leamy LJ, Pomp D, Eisen EJ, Cheverud JM. Quantitative trait loci for directional but not fluctuating asymmetry of mandibular characters in mice. Genet Res. 2000;76:27–40. doi: 10.1017/s0016672300004559. [DOI] [PubMed] [Google Scholar]
- 57.Klingenberg CP, Leamy LJ, Routman EJ, Cheverud JM. Genetic architecture of mandible shape in mice: effects of quantitative trait loci analyzed by geometric morphometrics. Genetics. 2001;157:785–802. doi: 10.1093/genetics/157.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishimura I, Drake TA, Lusis AJ, Lyons KM, Nadeau JH, Zernik J. ENU large-scale mutagenesis and quantitative trait linkage (QTL) analysis in mice: novel technologies for searching polygenetic determinants of craniofacial abnormalities. Crit Rev Oral Biol Med. 2003;14:320–30. doi: 10.1177/154411130301400503. [DOI] [PubMed] [Google Scholar]
- 59.Oh J, Wang CJ, Poole M, et al. A genome segment on mouse chromosome 12 determines maxillary growth. J Dent Res. 2007;86:1203–6. doi: 10.1177/154405910708601212. [DOI] [PubMed] [Google Scholar]
- 60.Haworth K, Breen M, Binns M, Hopkinson DA, Edwards YH. The canine homeobox gene MSX2: sequence, chromosome assignment and genetic analysis in dogs of different breeds. Anim Genet. 2001;32:32–6. doi: 10.1046/j.1365-2052.2001.00702.x. [DOI] [PubMed] [Google Scholar]
- 61.Haworth KE, Islam I, Breen M, et al. Canine TCOF1; cloning, chromosome assignment and genetic analysis in dogs with different head types. Mamm Genome. 2001;12:622–9. doi: 10.1007/s00335-001-3011-0. [DOI] [PubMed] [Google Scholar]
- 62.Haworth KE, Healy C, McGonnell IM, Binns M, Sharpe PT. Characterization of the genomic canine Fgf8 locus and screen for genetic variants in 4 dogs with different face types. DNA Seq. 2007;18:209–19. doi: 10.1080/10425170600986654. [DOI] [PubMed] [Google Scholar]
- 63.Fondon JW, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–63. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paternoster L, Zhurov AI, Toma AM, Kemp JP, St Pourcain B, Timpson NJ, McMahon G, McArdle W, Ring SM, Smith GD, Richmond S, Evans DM. Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am J Hum Genet. 2012;90:478–85. doi: 10.1016/j.ajhg.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu F, van der Lijn F, Schurmann C, Zhu G, Chakravarty MM, Hysi PG, Wollstein A, Lao O, de Bruijne M, Ikram MA, van der Lugt A, Rivadeneira F, Uitterlinden AG, Hofman A, Niessen WJ, Homuth G, de Zubicaray G, McMahon KL, Thompson PM, Daboul A, Puls R, Hegenscheid K, Bevan L, Pausova Z, Medland SE, Montgomery GW, Wright MJ, Wicking C, Boehringer S, Spector TD, Paus T, Martin NG, Biffar R, Kayser M. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 2012;8:e1002932. doi: 10.1371/journal.pgen.1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]