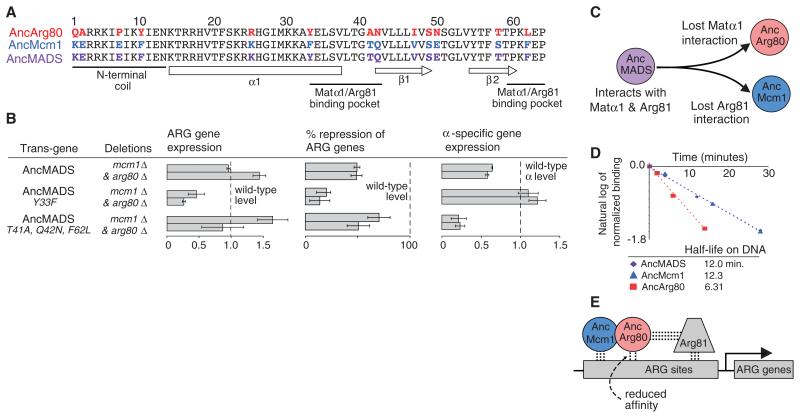
Fig. 3. Divergence in cofactor and DNA-binding following gene duplication of ancestral MADS-box proteins.
(A) Alignment of the N-terminal 63 amino acids of the MADS-box domain with residues that changed identity between AncMcm1, AncArg80, and AncMADS in color (24). α1 denotes a long α helix; β1 and β2 signify an antiparallel β sheet. (B) Gene expression profiling to determine the impact of mutants on the function of preduplication AncMADS protein in S. cerevisiae. Gene expression quantified using NanoString. Panel 1: MADS-box activated ARG genes; row 1, CAR1; row 2, CAR2. Panel 2: MADS-box repressed ARG genes; row 1, ARG3; row 2, ARG5,6. Panel 3: MADS-box activated mating genes (α-specific genes); row 1, SAG1; row 2, MFa1. Mean and standard error (indicated by error bars) were determined from three replicates. (C) After duplication, AncArg80 lost the ability to form a strong interaction with Matα1, and AncMcm1 lost the ability to form a strong interaction with Arg81. These losses destroyed the abilities of AncArg80 and AncMcm1 to regulate α-specific genes and ARG genes, respectively. (D) Half-lives of MADS-box ancestors on the S. cerevisiae CAR2 cis-regulatory sequence (see supplementary materials and methods). Saturating levels of unlabeled DNA were added at time point zero. (E) After the duplication of AncMADS, α-specific genes are regulated by a homodimer of AncMcm1, whereas ARG genes are regulated by a heterodimer of AncMcm1 and AncArg80 due to the reduced affinity of AncArg80 for DNA.