Abstract
Sumoylation, a post-translational protein modification by small ubiquitin-like modifier (SUMO), has been implicated in many stress responses. Here we analyzed the potential role of sumoylation in osmo-response in yeast. We find that osmotic stress induces rapid accumulation of sumoylated species in normal yeast cells. Interestingly, disruption of MAP kinase Hog1 leads to a much higher level of accumulation of sumoylated conjugates that are independent of new protein synthesis. We also find that the accumulation of sumoylated species is dependent on a SUMO ligase Siz1. Notably, overexpression of SIZ1 in HOG1-disruption mutants (hog1Δ) but not in wild type cells leads to a markedly increased and prolonged accumulation of sumoylated species. Examination of osmo-tolerance of yeast mutants that display either an increase or a decrease in the global sumoylation level revealed an inverse relationship between accumulation of sumoylated conjugates and osmo-tolerance. Further investigation has shown that many of the sumoylated species induced by hyperosmotic stress are actually poly-sumoylated. Together, these findings indicate that abnormal accumulation of poly-sumoylated conjugates is harmful for osmo-tolerance in yeast, and suggest that Hog1 promotes adaptation to hyperosmotic stress partially via regulation of global sumoylation level.
Introduction
Hog1 is a yeast mitogen-activated protein kinase (MAP kinase, or MAPK) that is required for cell survival in response to increased osmolarity in the environment [1], [2]. Yeast cells can sense osmolarity changes via one of two transmembrane proteins, Sho1 and Sln1, which subsequently triggers a cascade of enzymatic events leading to the activation of Hog1 [1], [3]. Hog1 appears to be necessary for changes elicited by high osmolarity stimulation that are required for cells to adapt and survive in such conditions, as evidenced by the inability of the hog1Δ cells to grow in the presence of high osmolarity [4]. The mechanisms by which Hog1 promotes osmo-tolerance are under intensive investigation. It was thought that Hog1 exerts its function mainly via phosphorylating nuclear transcriptional regulators such as Hot1 to modulate gene expression [5], [6]. Notably, an elegant subsequent work suggested that transcriptional regulation may not be the key mechanism by which Hog1 promotes osmo-tolerance, as preventing nuclear entrance of Hog1, a necessary step for Hog1 to modulate gene transcription, does not affect the ability of Hog1 to promote osmo-tolerance [7]. Thus, it appears that Hog1 may exert its essential role in osmo-adaptation via additional mechanisms, such as controlling the nuclear export of mRNA upon stress [8].
Post-translational modification of proteins by small ubiquitin-like modifier (SUMO) has emerged as a common mechanism for cell regulation [9]. Similar to the process of ubiquitination, SUMO is conjugated to a lysine residue on substrate via the action of E1 SUMO-activating enzyme, E2 SUMO-conjugating enzyme, and in certain cases E3 SUMO-ligating enzyme [9]. In yeast, Ubc9 is the only E2 enzyme for sumoylation, while Siz1, Siz2 and Mms21 are three known E3 ligases for sumoylation [10]–[12]. Just as ubiquitin can form poly-ubiquitin chains, SUMO can also be conjugated to other SUMO to form chains. Three lysine residues at the N-terminal of yeast SUMO (i.e., K11, K15, and K19) are the sites for the formation of poly-SUMO chain in yeast [13]. Sumoylation is also a reversible modification. In yeast, Ulp1 and Ulp2 are the two known de-sumoylating enzymes [14], [15]. It is believed that Ulp1 is responsible for de-conjugating solitary SUMO proteins while Ulp2 is responsible for disassembling poly-SUMO chains [13].
Sumoylation has been implicated in stress responses in a variety of systems including yeast, plant seedlings and mammalian cells [16]–[19]. Various stress conditions, such as oxidative stress, ethanol stress, osmotic stress as well as heat shock, can drastically increase the accumulation of sumoylated conjugates [16]–[18]. However, the underlying molecular mechanism and whether sumoylation is critical for stress adaptation are largely unclear. Work in plants demonstrated a clear correlation between accumulation of SUMO1/2-conjugated proteins and impairment in salt-tolerance [17]. This suggests that accumulation of sumoylated proteins to an abnormal level could be detrimental to stress adaptation. Therefore, mechanisms must exist to protect cells under osmotic stress by preventing abnormal accumulation of harmful SUMO-conjugates.
In this study, we examined the involvement of sumoylation in the yeast response to osmotic stress. We show that osmotic stress induces rapid accumulation of sumoylated conjugates but the level of accumulation is limited by a stress-activated MAP kinase Hog1. We find Siz1 but not Siz2 is mainly responsible for the accumulation of osmo-induced SUMO conjugates. Interestingly, overexpression of the SIZ1gene leads to a drastically increased and prolonged accumulation of osmo-induced sumoylated species in the hog1Δ mutant. Consistent with the notion that abnormal accumulation of sumoylated conjugates is harmful for stress tolerance, we find overexpression of SIZ1 exacerbates the sensitivity of the hog1Δ cells to hyperosmotic stress. Thus, our analysis reveals a new mechanism by which Hog1 promotes adaptation to osmotic stress, namely, via limiting the accumulation of harmful sumoylated conjugates induced by such stress.
Materials and Methods
Strains and Plasmids
Standard methods for the growth, maintenance, and transformation of yeast and bacteria and for the manipulation of DNA were used throughout. The yeast S. cerevisiae strains used in this study are BY4741 (MAT a his3Δ leu2Δmet15Δura3Δ), BY4741-derived mutants lacking HOG1, SIZ1, and SIZ2 (Research Genetics, Huntsville, AL), BY4741-derived mutant carrying a catalytic inactive allele of HOG1 (hog1K52R) (from Stephen Parnell at University of Kansas), BY4741-derived mutant lacking both HOG1 and SIZ1 (BY4741 hog1::LEU2 siz1::KanMX, this work), W303 strain MHY500 (MAT a his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1),MHY500-derived mutants harboring temperature-sensitive allele of ubc9-1 (MAT a his3-Δ200 leu2-3,112::LEU2::ubc9-1 ura3-52 lys2-801 trp1-1 ubc9Δ::TRP1) and ulp1-333 (MAT a his3-Δ200 leu2-3,112::LEU2::ulp1-333 ura3-52 lys2-801 trp1-1 ulp1Δ1::his3::URA3), MHY500-derived mutants lacking ULP2 (ulp2-Δ1::HIS3) from Mark Hochstrasser at Yale [15], JD-52 strain (MAT a trp1-Δ1 ura3-52 his3-Δ200 leu2-3,112 lys2-801) and JD-52-derived mutants IS30 (ulp2Δ::URA3), GBY1 (MAT a smt3-R11,15,19::TRP1), GBY7 (MAT a smt3-R11,15,19::TRP1 ulp2Δ::URA3) from Erica Johnson at Thomas Jefferson University [13], and IS30-derived mutants lacking both ULP2 and HOG1 (ulp2Δ::URA3 hog1Δ::LEU2, this work). Plasmid overexpressing SIZ1, i.e., BG1805-GAL-SIZ1-TAP was purchased from OpenBiosystems.
Growth, Phosphorylation, and Sumoylation Bioassays
Growth assays were conducted as described previously [20]. Phosphorylation of Hog1 was monitored by immunoblotting of whole cell extracts, using antibodies that recognize dually phosphorylated p38 [21]. For all the immunoblotting analysis, mid-log cell cultures were grown on appropriate medium, treated or not treated with 1 M sorbitol for indicated length of time. Growth was stopped by the addition of 10 mM NaN3 and transfer to an ice bath. Cells were washed and proteins were extracted via trichloroacetic precipitation, following procedures described previously [22]. Whole cell extracts were resuspended in boiling SDS-PAGE sample buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 1% 2-mercaptoethanol, 0.0005% bromphenol blue) for 5 min. Following SDS-polyacrylamide gel electrophoresis and transfer to nitrocellulose, the membrane was probed with antibodies to phosphor-p38 at 1∶1,000 (from Cell Signaling), and SUMO at 1∶10,000 (from Stefan Jentsch, Max Planck Institute of Biochemistry, Germany). Immunoreactive species were visualized by enhanced chemiluminescence detection (Pierce) of horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad) or anti-goat IgG (Santa Cruz Biotechnology). Specificity of detection was established using hog1Δ, and ubc9-1 cell extracts as negative controls. Note that an efficient detection of very high molecular weight species (on the very top of the gel) accumulated in the ulp2Δ mutant requires anti-SUMO antibody to be very fresh. All experiments have been repeated two or three times.
Results
Hog1 is a MAP kinase in yeast that is primarily responsible for eliciting responses that ensure proper adaptation and survival of cells to hyperosmotic stress [1]. We were interested in understanding mechanisms by which Hog1 promotes adaptation to high osmolarity. Our objective here is to investigate whether Hog1 is involved in the regulation of global sumoylation. Previously, it has been shown in plants that salt treatment leads to accumulation of sumoylated proteins [17], [19]. Intriguingly, the degree of accumulation of sumoylated proteins seems to inversely correlate with the capability of the plant seedlings to tolerate salt stress [17]. This suggests that abnormal accumulation of sumoylated proteins could be detrimental to cells and mechanisms must exist to limit the accumulation of sumoylated proteins.
We hypothesize that signaling pathway evoked in response to hyperosmotic stress could be responsible for preventing accumulation of sumoylated proteins to a level that is detrimental to the cells. To test this hypothesis in yeast, we first determined whether hyperosmotic stress also induces accumulation of sumoylated conjugates in yeast. For this purpose, we examined the level of sumoylated conjugates in cells that were treated with 1 M sorbitol over time. Indeed, hyperosmotic stress induced a transient but substantial accumulation of sumoylated conjugates at the time point of 15 minutes, followed by a gradual decline of these conjugates ( Fig. 1 ). Interestingly, the decline of sumoylated conjugates occurred right after the maximal activation of Hog1, a MAP kinase that is responsible for normal osmo-tolerance in yeast, as detected by anti-phospho-p38 that recognizes dually phosphorylated and thus activated Hog1 ( Fig. 1 , lower panel) [21], [23]. To determine whether Hog1 has any inhibitory role on the accumulation of sumoylated conjugates induced by hyperosmotic stress, we conducted similar experiment in a hog1Δ mutant. Remarkably, a much higher level of sumoylated conjugates is accumulated in the hog1Δ mutant, as compared to wild type cells ( Fig. 1 ). This suggests that Hog1 indeed has a role in limiting the accumulation of sumoylated conjugates triggered by hyperosmotic stress.
Figure 1. A MAP kinase Hog1 limits the accumulation of sumoylated conjugates induced by hyperosmotic stress.
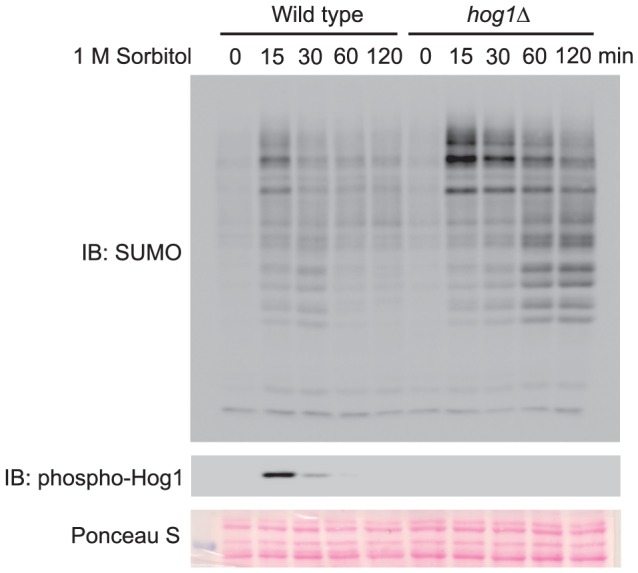
Whole cell extracts were prepared from wild type and hog1Δ mutants treated with 1 M sorbitol for the indicated time, resolved by 8% SDS-PAGE, and probed with anti-SUMO (top panel) or anti-phospho-Hog1 (middle panel) antibodies. The specificity of SUMO antibody was established by previous work and was further confirmed by cell extracts from SUMO E2-defective mutant cells (data not shown). The equal loading of each lane was confirmed by Ponceau S staining (bottom panel). IB, immunoblotting.
To determine whether the kinase activity of Hog1 is required in this process, we examined the behavior of hog1K52R, a catalytically inactive mutant of Hog1. In this mutant, a lysine residue of Hog1 that is responsible for binding ATP has been changed to an arginine. Consequently, this mutant can be phosphorylated by its upstream kinase Pbs2 but lacks the ability to phosphorylate other proteins. As reported previously, in response to sorbitol stimulation, the hog1K52R mutant displayed prolonged phosphorylation, compared to wild type, as detected by immunoblotting with anti-phospho-p38 antibody ( Fig. 2 , middle panel) [3], [23]. Similar to the hog1Δ mutants, a higher level of sumoylated conjugates is accumulated in the hog1K52R mutants, especially in the later time points ( Fig. 2 , upper panel). Thus, inhibiting the accumulation of osmo-triggered sumoylation requires the kinase activity of Hog1.
Figure 2. Catalytic activity of Hog1 is required for its inhibition of global sumoylation.
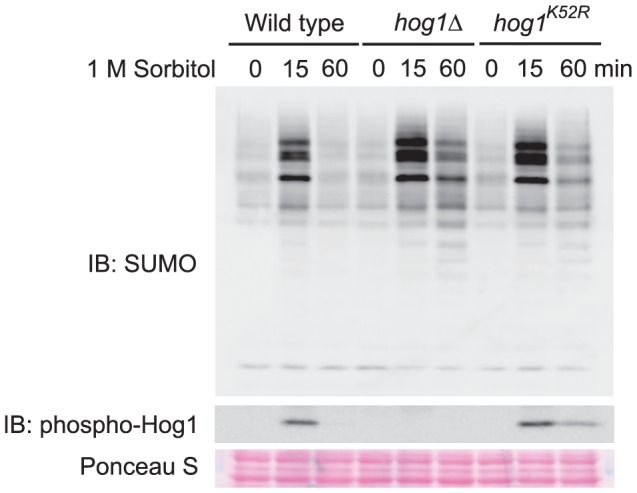
Whole cell extracts were prepared from wild type, hog1Δmutants, and hog1K52R mutants treated with 1 M sorbitol for the indicated time, resolved by 8% SDS-PAGE, and probed with anti-SUMO (top panel) or anti-phospho-Hog1 (middle panel) antibodies. The equal loading of each lane was confirmed by Ponceau S staining (bottom panel). IB, immunoblotting.
Many effects exerted by Hog1 are mediated via activation of its downstream transcription factors, which consequently leads to new gene transcription and new protein synthesis. It is possible that regulation of global sumoylation by Hog1 is mediated by its downstream transcription factors. In that case, we expect that new protein synthesis might be required for the inhibitory effect of Hog1. To test this, we repeated the experiments in wild type and the hog1Δ mutants, with or without the addition of protein synthesis inhibitor cycloheximide. As shown in Fig. 3 , cycloheximide (CHX) treatment has nearly no effect on changes in the global sumoylation triggered by hyperosmotic stress, in either wild type or the hog1Δ mutants, especially in the later time points (i.e., 60 min). Thus, the effect of Hog1 on limiting the accumulation of osmo-induced sumoylation does not require new protein synthesis, therefore excluding the possibility that the effect is mediated via transcription factors controlled by Hog1.
Figure 3. New protein synthesis is NOT required for Hog1 to prevent the accumulation of sumoylated conjugates induced by hyperosmotic stress.
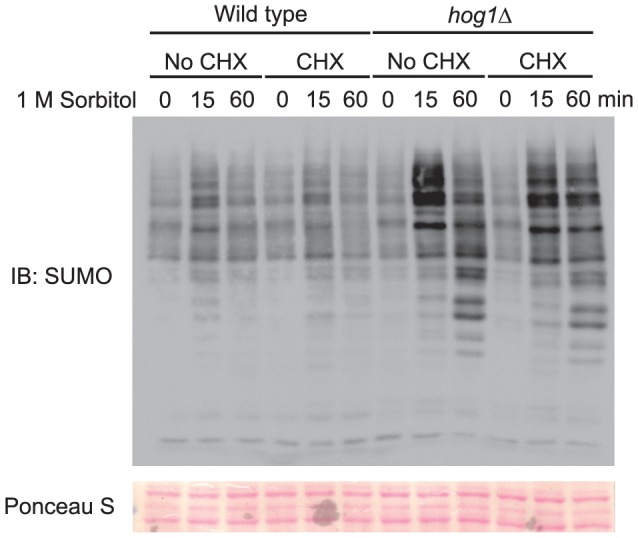
Whole cell extracts were prepared from wild type and hog1Δmutants treated with protein synthesis inhibitor cycloheximide (CHX) to stop new protein synthesis, followed by 1 M sorbitol for the indicated time, resolved by 8% SDS-PAGE, and probed with anti-SUMO antibodies. The equal loading of each lane was confirmed by Ponceau S staining. IB, immunoblotting.
Next, we consider the possibility that Hog1 regulates global sumoylation via modulating the activity of enzymes in the sumoylation pathway. To test this possibility, we sought to identify the ligase that is responsible for the observed accumulation of sumoylated conjugates. Siz1 and Siz2 are two well characterized E3 ligases that catalyze protein sumoylation in yeast [11]. To determine if they are responsible for hyperosmotic stress-induced sumoylation, we examined the sumoylation status of the siz1Δ and siz2Δ mutants, with or without hyperosmotic stress. As shown in Fig. 4 , sumoylated species observed in wild type cells were largely absent in the siz1Δ but not the siz2Δ mutants. Thus Siz1 appears to be the main ligase that is responsible for the formation of sumoylation conjugates. The residual signals detected in the siz1Δ mutants indicate that additional ligase may also have a minor role in this process.
Figure 4. SUMO ligase Siz1 but not Siz2 is required for hyperosmotic stress-induced accumulation of sumoylated conjugates.
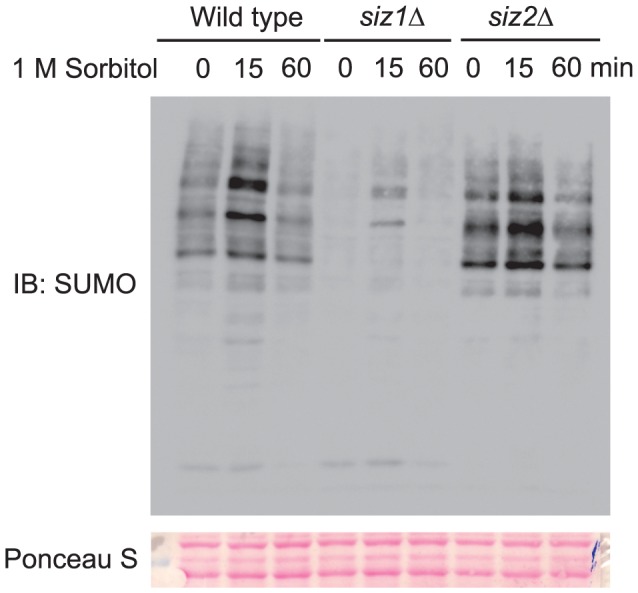
Whole cell extracts were prepared from wild type, siz1Δ, and siz2Δ mutants treated with 1 M sorbitol for the indicated time, resolved by 8% SDS-PAGE, and probed with anti-SUMO antibodies. The equal loading of each lane was confirmed by Ponceau S staining. IB, immunoblotting.
Having identified Siz1 as the main ligase for the observed global sumoylation, we then proceeded to examine whether overexpression of SIZ1 has any effect on the accumulation of sumoylated conjugates in hyperosmotic stressed cells. As shown in Fig. 5 , overexpression of SIZ1 in wild type cells has negligible effect on sumoylation (compare Fig. 5 and Fig. 1 ). However, overexpression of SIZ1 in the hog1Δ cells leads to a significantly enhanced and prolonged accumulation of SUMO-conjugates ( Fig. 5 ). The drastically different outcomes of overexpressing SIZ1 in wild type versus the hog1Δ mutants suggest that Hog1 has an inhibitory role on the activity of Siz1. As such, overexpressed Siz1 is rendered largely inactive in wild type cells. In contrast, the inhibitory effect of Hog1 on overexpressed Siz1 is relieved in the hog1Δ mutants, leading to an increased accumulation of SUMO-conjugates in osmotically stressed cells.
Figure 5. Overexpression of SIZ1 in the hog1Δ mutants but not in the wild type increases hyperosmotic stress-induced sumoylation.
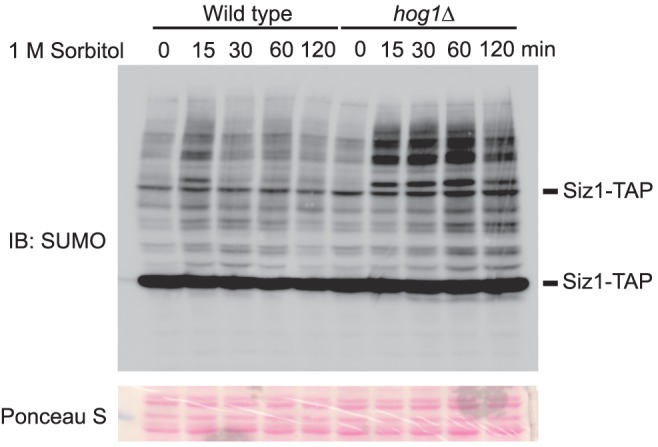
Wild type or the hog1Δ cells were transformed with plasmids that overexpress TAP-tagged Siz1 (i.e., Siz1-TAP). Cells were grown to mid-log phase, treated with 1 M sorbitol for the indicated time, and harvested. Whole cell extracts were prepared and resolved by 8% SDS-PAGE, and probed with anti-SUMO antibodies. Note that TAP tag contains IgG binding motif, and thus Siz1-TAP was also recognized by SUMO antibody. For unknown reason, two bands with different sizes of Siz1-TAP were detected, and these two bands are absent in cells transformed with an empty vector.
Having identified situations that either diminish (i.e., siz1Δ mutants) or enhance (i.e., hog1Δ mutants, and overexpressing Siz1 in the hog1Δ mutants) osmo-induced accumulation of SUMO-conjugates, we then examined whether there is any correlation between the level of SUMO-conjugates and the ability of the cells to adapt to hyperosmotic stress. For this purpose, we compared the growth of these different cells with or without the addition of 1 M sorbitol in the plates. As shown in Fig. 6a , both wild type and the siz1Δ mutants grew equally well with or without the addition of 1 M sorbitol, suggesting that accumulation of SUMO-conjugates triggered by hyperosmotic stress is not required for normal adaptation. As expected [2], disrupting Hog1 (i.e., hog1Δ mutants) renders the cells sensitive to hyperosmotic stress ( Fig. 6b ). Interestingly, the osmo-sensitivity of the hog1Δ mutants is significantly exacerbated by overexpressing the SIZ1 gene. This synergistic effect is especially evident when a mild hyperosmotic stress (e.g., 0.5 M sorbitol) was applied ( Fig. 6b ). These data suggest an inverse relationship between the level of SUMO-conjugates and the tolerance of cells to hyperosmotic stress.
Figure 6. Overexpression of SIZ1 enhances osmo-sensitivity of hog1Δ mutants.
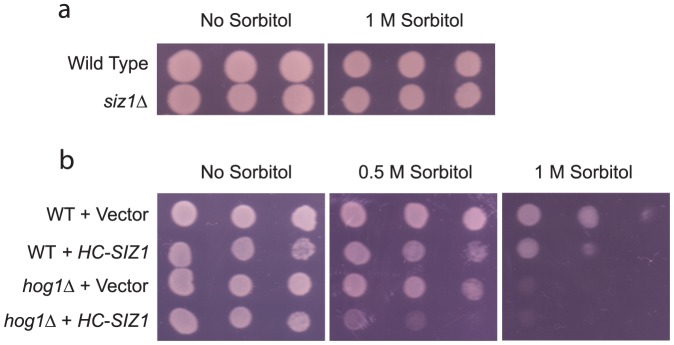
a, Osmo-sensitivity of wild type and siz1Δ mutants was measured by comparing the growth of serial diluted exponentially growing yeast cells on YPD plates with or without the addition of 1 M sorbitol. b, wild type (WT) and hog1Δ mutants were transformed with either URA3-marked plasmids that overexpress SIZ1 (i.e., HC-SIZ1) or URA3-marked empty vector. The osmo-sensitivity of the cells was measured by comparing the growth of serial diluted exponential cells on SCG-URA plates with or without the addition of various concentrations of sorbitol (0.5 M and 1 M, respectively).
To further confirm a potential correlation between accumulation of SUMO-conjugates and osmo-intolerance, we also compared the growth of mutants lacking a functional Ubc9 (the sole E2 for sumoylation), Ulp1 (desumoylating enzyme), or Ulp2 (another desumoylating enzyme), with or without hyperosmotic stress. It has been shown previously that lacking a functional Ubc9 abolishes nearly all sumoylation events [10], while lacking a functional Ulp1 or Ulp2 leads to accumulation of SUMO-conjugates [14], [15]. As shown in Fig. 7a , similar to the siz1Δ mutants, lacking a functional Ubc9 has nearly no effect on the cell growth in the presence of 1 M sorbitol. Lacking a functional desumoylating enzyme Ulp1 has a modest effect on cell growth in the presence of 1 M sorbitol, while disrupting Ulp2 has a drastic effect. These results are consistent with our notion that sumoylation per se is not required for proper adaptation to hyperosmotic stress but abnormal accumulation of sumoylated species hinders the osmo-adaptation response.
Figure 7. An accumulation of poly-sumoylated conjugates renders the ulp2Δ mutants more sensitive to hyperosmotic stress.
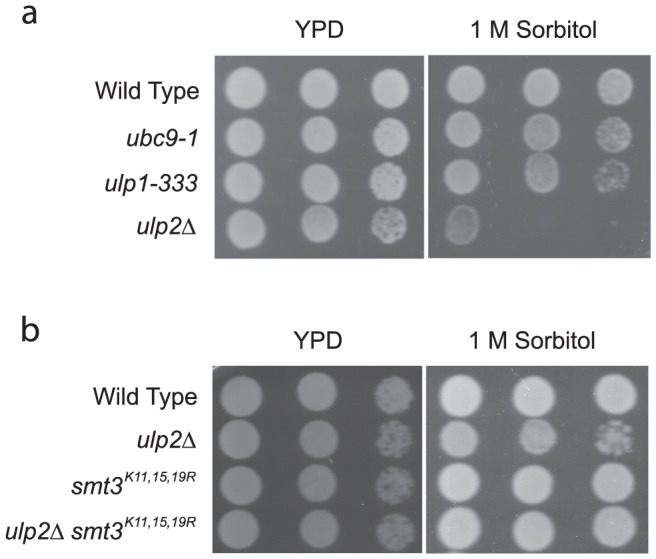
a, osmo-sensitivity of wild type (MHY500) and MHY500-derived mutants ubc9-1, ulp1-333 and ulp2Δ, was measured by comparing the growth of serial diluted exponentially growing yeast cells on YPD plates with or without the addition of 1 M sorbitol. b, osmo-sensitivity of wild type (JD-52) and JD-52 derived mutants either accumulating poly-sumoylated species (ulp2Δ) or incapable of forming poly-sumoylated species (smt3K11,15,19R and ulp2Δ/smt3K11,15,19R) was similarly measured.
It has been shown that Ulp1 and Ulp2 have distinct substrates. Specifically, disrupting Ulp1 leads to accumulation of mono-SUMO conjugated to substrates while disrupting Ulp2 leads to accumulation of poly-SUMO conjugates [13]. To test whether accumulation of poly-sumoylated species is responsible for the severe osmo-sensitivity of the ulp2Δ mutant, we examined whether blocking formation of poly-sumoylated species in the mutant can rescue its osmo-sensitivity. Poly-sumoylation can be prevented by mutating three lysine residues on SUMO that are necessary for chain formation (i.e., K11, K15, and K19) [24]. Accordingly, we compared the osmo-sensitivity of the ulp2Δ mutant and the ulp2Δsmt3K11, 15, 19R double mutant. As shown in Fig. 7b , the ulp2Δsmt3K11, 15, 19R double mutant grew much better than the ulp2Δ mutant in the presence of high osmolarity, suggesting that abnormal accumulation of poly-sumoylated species significantly contributes to the severe osmo-sensitivity of the ulp2Δ mutant.
We then examined whether hyperosmotic stress induces accumulation of poly-sumoylated species. For this purpose, we analyzed the global sumoylation level of wild type cells versus various mutants that either expresses mutant allele of SUMO or lacks ULP2, or both. As shown in Fig. 8 , high molecular weight species were accumulated in the ulp2Δ mutants and mutating the three lysine residues to arginine as indicated drastically diminished the SUMO signal. Notably, hyperosmotic stress-induced change in global sumoylation is most prominent in wild type cells and such hyperosmotic stress-induced change is less apparent in the mutants (ulp2Δ, smt3K11, 15, 19R and ulp2Δsmt3K11, 15, 19R). These data indicate that sumoylated species induced by hyperosmotic stress are largely poly-sumoylated conjugates.
Figure 8. Hyperosmotic stress induces an accumulation of poly-sumoylated conjugates.
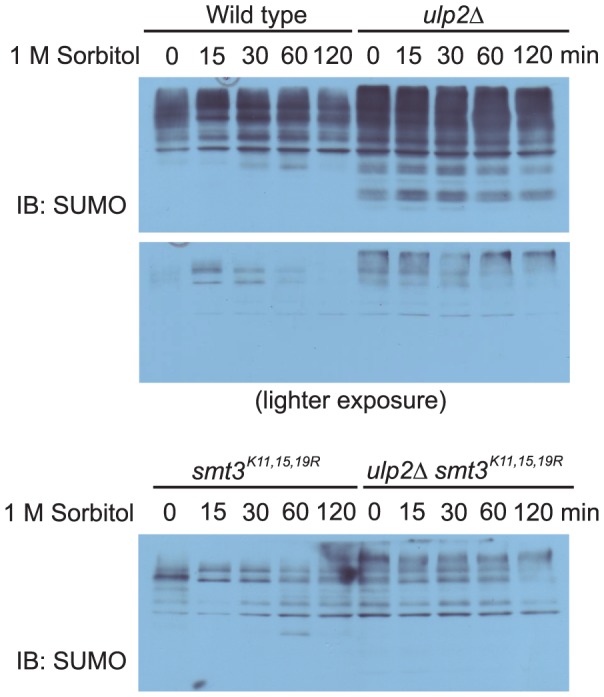
Whole cell extracts were prepared from wild type (JD-52) and JD-52 derived mutants either accumulating poly-sumoylated species (ulp2Δ) or incapable of forming poly-sumoylated species (smt3K11,15,19R and ulp2Δ/smt3K11,15,19R) treated with 1 M sorbitol for the indicated time, resolved by 8% SDS-PAGE, and probed with anti-SUMO antibodies. IB, immunoblotting. Note that an efficient detection of the very high molecular weight species accumulated in the ulp2Δ mutants requires the antibody (anti-SUMO) to be very fresh.
Finally, we examined whether Siz1 and/or Ulp2 are required for Hog1 to regulate hyperosmolarity-induced sumoylation. For this purpose, we constructed double mutants (hog1Δsiz1Δ and hog1Δ ulp2Δ) and examined whether deleting either SIZ1 or ULP2 abolishes the enhanced accumulation of sumoyalted species in the hog1Δ cells. As shown in Fig. 9a , the marked accumulation of poly-sumoylated species observed in the hog1Δcells in response to hyperosmotic stress is substantially diminished in the hog1Δsiz1Δ double mutants. Thus, Siz1 is partially responsible for the further enhanced accumulation of poly-sumoylated species in the hog1Δcells. Deleting ULP2 also diminishes the level of accumulation of poly-sumoylated species in the hog1Δ cells ( Fig. 9b ). Together, these results indicate that both Siz1 and Ulp2 are required for Hog1 to regulate the optimal level of hyperosmolarity-induced poly-sumoylation.
Figure 9. Both Siz1 and Ulp2 are required for Hog1 to limit accumulation of sumoylated conjugates induced by hyperosmotic stress.
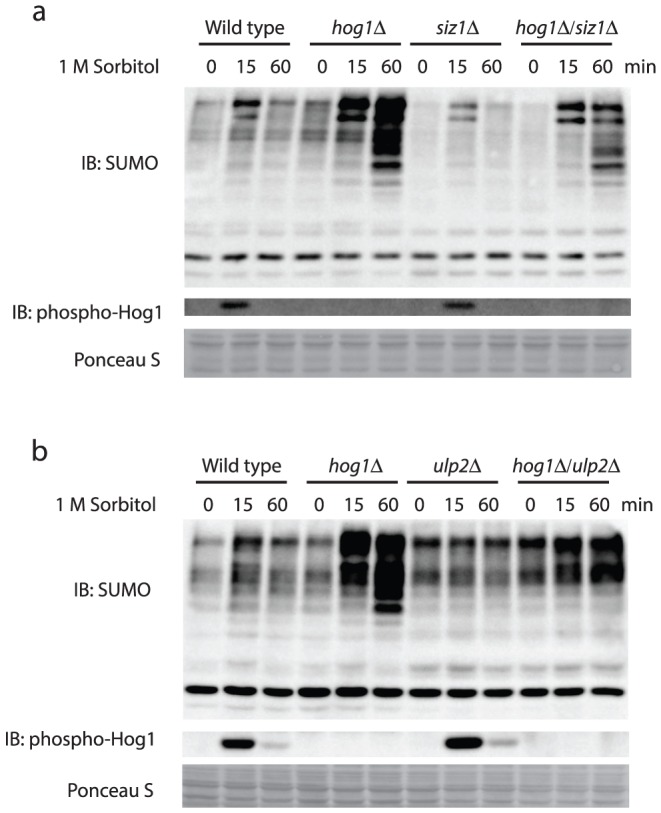
a & b, Whole cell extracts were prepared from wild type, hog1Δ, siz1Δ, hog1Δ/siz1Δ, ulp2Δ, and hog1Δ/ulp2Δ treated with 1 M sorbitol for the indicated time, resolved by 8% SDS-PAGE, and probed with anti-SUMO antibodies. IB, immunoblotting.
Discussion
Most cells have the capacity to cope with environmental stresses such as changes in osmolarity. In yeast, successful adaptation to hyperosmotic stress requires the action of Hog1, a stress-activated protein kinase. In this study, we provide evidence that Hog1 promotes adaptation to high osmolarity partially via preventing and/or removal of the harmful accumulation of sumoylated species. We find that hyperosmotic stress induces a transient increase in the level of global sumoylation, but blocking sumoylation itself has no or negligible effects on long term osmo-adaptation, indicating that sumoylation per se does not play any apparent positive roles in promoting adaptations to hyperosmotic stress. However, aberrant accumulation of such species renders cells incapable of coping with hyperosmotic stress, and Hog1 prevents this catastrophic event from happening by limiting the accumulation of such harmful sumoylated species. In addition, we find the sumoylated species induced by hyperosmotic stress are largely poly-sumoylated. Together, these findings suggest that abnormal accumulation of poly-sumoylated species is harmful for osmo-tolerance, and that Hog1 promotes adaptation to high osmolarity partially via regulating the level of global poly-sumoylation.
It is unclear why abnormal accumulation of poly-sumoylated species is harmful for osmo-tolerance. One well-established function of poly-sumoylation is serving as an alternative signal that targets substrate proteins for proteasomal degradation. In this SUMO-targeted protein degradation pathway, poly-sumoylated substrates are recognized by the E3 ubiquitin ligase Slx5/Slx8, which contains SUMO-interacting motif (SIM), and become ubiquitinated and subsequently degraded in the proteasome [25], [26]. In theory, it appears possible that abnormal accumulation of poly-sumoylated species could alter the half-lives of certain proteins that are crucial for appropriate osmo-adaptation. However, neither slx5Δnor slx8Δcells display any altered sensitivity to hyperosmotic stress (data not shown). In fact, a survey of a library of deletion mutants that lack each of the known or putative E3 ubiquitin ligases in yeast [27] did not reveal any that displays an altered sensitivity to hyperosmotic stress (data not shown). Therefore, poly-sumoylation likely affects osmo-adaptation independent of targeting proteins for degradation.
Another possibility is that abnormal accumulation of poly-sumoylated species may hinder appropriate gene expression induced by hyperosmotic stress that is required for adaptation. When challenged with hyperosmotic stress, yeast cells initially repress overall gene expression by altering chromatin structure and releasing most regulating proteins from chromatin [28]. This transient gene repression phase (lasting less than 5 minutes) is followed by the induction of hundreds of genes, many of which are under the control of Hog1-activated transcription factors such as Hot1 [29]–[31]. A recent study indicated that SUMO chains (possibly via poly-sumoylation of certain protein substrates) are important for the maintenance of chromatin structure and transcriptional repression of stress-induced genes [32]. It is conceivable that rapid up-regulation of poly-sumoylation could initially help pausing global gene expression as an immediate response to hyperosmotic stress; however, abnormal accumulation (as seen in the hog1Δ cells) could have a detrimental effect on the derepression of numerous stress responsive genes, thereby leading to a defect in long-term adaptation to hyperosmotic stress. The transient nature of the poly-sumoylation response points to an effort by the cell to limit extended global SUMO conjugation; furthermore, the carefully timed nature of this response also lends support to the fact that prolonged accumulation of poly-sumoylated species is detrimental to the cell's adaptation to osmotic-stress.
Our data clearly showed that the activity of Hog1 is required for the maintenance of normal poly-sumoylation level induced by hyperosmotic stress. What could be the molecular mechanism by which Hog1 regulates poly-sumoylation? Our findings that the Hog1 effect is independent of new protein synthesis suggest that the regulation is not mediated via activation of transcription factors such as Hot1 that are under the control of Hog1. One possibility is that Hog1 directly regulates the activity of enzymes in the sumoylation pathway such as Siz1 and Ulp2, given that both proteins are required for Hog1 to limit the level of poly-sumoylation. However, neither Siz1 nor Ulp2 contains a preferred Hog1 phosphorylation site (PXS/TP), and phos-tag analysis of these two proteins did not reveal any obvious Hog1-dependent mobility shift. Thus, it is unlikely that Hog1 regulates these two enzymes via direct phosphorylation. Our initial attempt to detect a potential interaction between Hog1 and these two enzymes (Siz1 and Ulp2) was also not successful, suggesting either the interaction is too weak or too transient for detection, or that Hog1 regulates the activity of those enzymes indirectly. Future studies will be directed to elucidate the molecular mechanisms by which Hog1 regulates sumoylation pathway.
Our genetic analysis indicates that deleting either SIZ1 or ULP2 can partially suppress the accumulation of sumoylated species in the hog1Δ mutants. It is easy to understand why deleting SIZ1 can suppress the accumulation of sumoylated species in the hog1Δ mutants because SIZ1 encodes a sumoylating enzyme that is responsible for hyperosmotic stress-triggered sumoylation. Why does deleting ULP2 also suppress the accumulation of sumoylated species in the hog1Δ cells, given that ULP2 encodes a desumoylating enzyme? One possibility is that Ulp2 may have a role in regulating the abundance of Siz1. A very recent report has shown that the abundance of Siz1 can be regulated by SUMO-targeted ubiquitination pathway [33]. In this pathway, poly-sumoylation of the substrate acts as a signal for its subsequent ubiquitination and degradation [26]. Given the role of Ulp2 in reversing poly-sumoylation, Ulp2 may desumoylate Siz1 and thereby rescue Siz1 from ubiquitination and degradation by the SUMO-targeted ubiquitination pathway. Consequently, deleting ULP2 may speed up the degradation of Siz1, which could result in suppressing the accumulation of sumoylated proteins in the hog1Δ mutants. This and other possibilities will be investigated in the future.
Acknowledgments
We thank Drs. Mark Hochstrasser, Erica Johnson and Stephen Parnell for generously providing strains, and Drs. Carsten Hoege, Stefan Jentsch, and Henrik Dohlman for kindly providing antibodies.
Funding Statement
This work was supported by a start-up fund from Saint Louis University and a National Institutes of Health (NIH) grant 1R15GM106330-01 (to Y.W.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chen RE, Thorner J (2007) Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1773: 1311–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC (1993) An osmosensing signal transduction pathway in yeast. Science 259: 1760–1763. [DOI] [PubMed] [Google Scholar]
- 3. Westfall PJ, Thorner J (2006) Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: use of an analog-sensitive HOG1 allele. Eukaryot Cell 5: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J (1996) Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol 16: 3637–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, et al. (1999) Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19: 5474–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rep M, Krantz M, Thevelein JM, Hohmann S (2000) The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem 275: 8290–8300. [DOI] [PubMed] [Google Scholar]
- 7. Westfall PJ, Patterson JC, Chen RE, Thorner J (2008) Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci U S A 105: 12212–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Regot S, de Nadal E, Rodriguez-Navarro S, Gonzalez-Novo A, Perez-Fernandez J, et al. (2013) The Hog1 Stress-activated Protein Kinase Targets Nucleoporins to Control mRNA Export upon Stress. J Biol Chem 288: 17384–17398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382. [DOI] [PubMed] [Google Scholar]
- 10. Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272: 26799–26802. [DOI] [PubMed] [Google Scholar]
- 11. Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744. [DOI] [PubMed] [Google Scholar]
- 12. Potts PR, Yu H (2005) Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 25: 7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bylebyl GR, Belichenko I, Johnson ES (2003) The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem 278: 44113–44120. [DOI] [PubMed] [Google Scholar]
- 14. Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398: 246–251. [DOI] [PubMed] [Google Scholar]
- 15. Li SJ, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20: 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou W, Ryan JJ, Zhou H (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279: 32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, et al. (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, et al. (2009) System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2: ra24. [DOI] [PubMed] [Google Scholar]
- 19. Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, et al. (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872. [DOI] [PubMed] [Google Scholar]
- 20. Wang Y, Dohlman HG (2006) Pheromone-regulated sumoylation of transcription factors that mediate the invasive to mating developmental switch in yeast. J Biol Chem 281: 1964–1969. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Abu Irqeba A, Ayalew M, Suntay K (2009) Sumoylation of transcription factor Tec1 regulates signaling of mitogen-activated protein kinase pathways in yeast. PLoS One 4: e7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG (2006) Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell 126: 191–203. [DOI] [PubMed] [Google Scholar]
- 23. Hao N, Zeng Y, Elston TC, Dohlman HG (2008) Control of MAPK specificity by feedback phosphorylation of shared adaptor protein Ste50. J Biol Chem 283: 33798–33802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Witowsky JA, Johnson GL (2003) Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J Biol Chem 278: 1403–1406. [DOI] [PubMed] [Google Scholar]
- 25. Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, et al. (2007) The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282: 34176–34184. [DOI] [PubMed] [Google Scholar]
- 26.Mullen JR, Brill SJ (2008) Activation of the SLX5-SLX8 ubiquitin ligase by poly-SUMO conjugates. J Biol Chem. [DOI] [PMC free article] [PubMed]
- 27. Ravid T, Hochstrasser M (2007) Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol 9: 422–427. [DOI] [PubMed] [Google Scholar]
- 28. Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, et al. (2006) The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell 23: 241–250. [DOI] [PubMed] [Google Scholar]
- 29. Nagiec MJ, Dohlman HG (2012) Checkpoints in a yeast differentiation pathway coordinate signaling during hyperosmotic stress. PLoS Genet 8: e1002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saito H, Tatebayashi K (2004) Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem (Tokyo) 136: 267–272. [DOI] [PubMed] [Google Scholar]
- 31. Saito H, Posas F (2012) Response to hyperosmotic stress. Genetics 192: 289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srikumar T, Lewicki MC, Costanzo M, Tkach JM, van Bakel H, et al. (2013) Global analysis of SUMO chain function reveals multiple roles in chromatin regulation. J Cell Biol 201: 145–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westerbeck JW, Pasupala N, Guillotte M, Szymanski E, Matson BC, et al. (2013) A SUMO-targeted ubiquitin ligase is involved in the degradation of the nuclear pool of the SUMO E3 ligase Siz1. Mol Biol Cell. [DOI] [PMC free article] [PubMed]


